4PL0
 
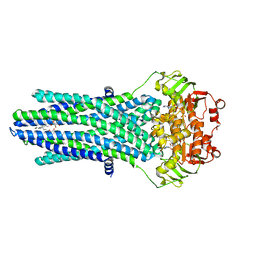 | |
2XVA
 
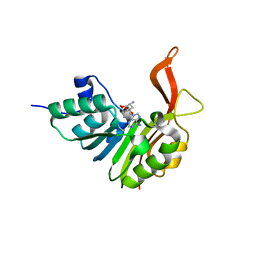 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with sinefungin | | Descriptor: | SINEFUNGIN, TELLURITE RESISTANCE PROTEIN TEHB, ZINC ION | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
2XVM
 
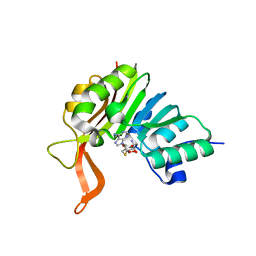 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TELLURITE RESISTANCE PROTEIN TEHB | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
5EG1
 
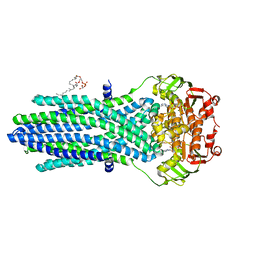 | | Antibacterial peptide ABC transporter McjD with a resolved lipid | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, ... | | Authors: | Choudhury, H.G, Beis, K. | | Deposit date: | 2015-10-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural and Functional Basis for Lipid Synergy on the Activity of the Antibacterial Peptide ABC Transporter McjD.
J.Biol.Chem., 291, 2016
|
|
5OFP
 
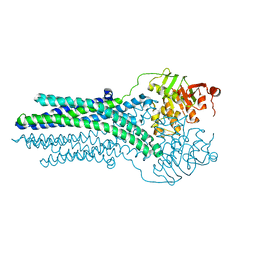 | |
4YCR
 
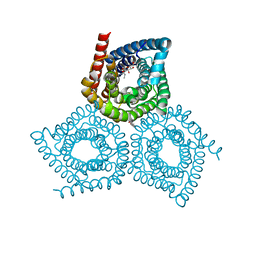 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | Descriptor: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | Authors: | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ID4
 
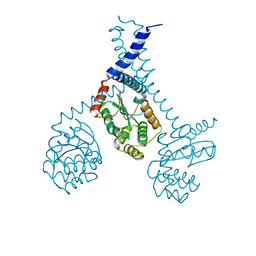 | |
5IDR
 
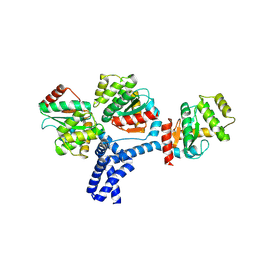 | |
6C29
 
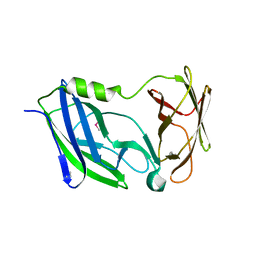 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | Descriptor: | Putative metal resistance protein | | Authors: | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | Deposit date: | 2018-01-07 | | Release date: | 2018-03-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
