1S5Z
 
 | | NDP kinase in complex with adenosine phosphonoacetic acid | | Descriptor: | ADENOSINE PHOSPHONOACETIC ACID, Nucleoside diphosphate kinase, cytosolic, ... | | Authors: | Chen, Y, Morera, S, Pasti, C, Angusti, A, Solaroli, N, Veron, M, Janin, J, Manfredini, S, Deville-Bonne, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adenosine phosphonoacetic acid is slowly metabolized by NDP kinase.
Med.Chem., 1, 2005
|
|
1UCN
 
 | | X-ray structure of human nucleoside diphosphate kinase A complexed with ADP at 2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chen, Y, Gallois-Montbrun, S, Schneider, B, Veron, M, Morera, S, Deville-Bonne, D, Janin, J. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide Binding to Nucleoside Diphosphate Kinases: X-ray Structure of Human NDPK-A in Complex with ADP and Comparison to Protein Kinases
J.Mol.Biol., 332, 2003
|
|
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
5BUS
 
 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in complex with AMP | | Descriptor: | 2-succinylbenzoate--CoA ligase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
5BUQ
 
 | | Unliganded Form of O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, Solved at 1.98 Angstroms | | Descriptor: | 2-succinylbenzoate--CoA ligase, ACETATE ION, CALCIUM ION | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
5BUR
 
 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in Complex with ATP and Magnesium Ion | | Descriptor: | 1,2-ETHANEDIOL, 2-succinylbenzoate--CoA ligase, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
3TOJ
 
 | | Structure of the SPRY domain of human Ash2L | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Chen, Y, Cao, F, Wan, B, Dou, Y, Lei, M. | | Deposit date: | 2011-09-05 | | Release date: | 2012-01-25 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30.
Cell Res., 22, 2012
|
|
5GTM
 
 | | Modified human MxA, nucleotide-free form | | Descriptor: | Interferon-induced GTP-binding protein Mx1 | | Authors: | Chen, Y, Gao, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Conformational dynamics of dynamin-like MxA revealed by single-molecule FRET
Nat Commun, 8, 2017
|
|
8UAQ
 
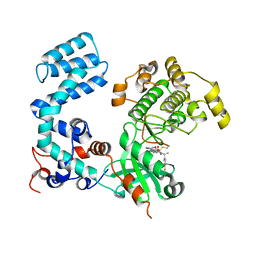 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 in Complex with GRL018-21 | | Descriptor: | (3Z)-N-[(1R)-1-(4-fluorophenyl)ethyl]-3-[(4-{[(2S)-2-(furan-2-yl)-2-hydroxyacetyl]amino}-3,5-dimethyl-1H-pyrrol-2-yl)methylidene]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
8UAP
 
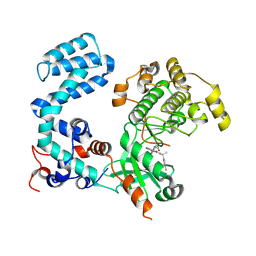 | | Crystal Structure of Human G Protein-Coupled Receptor Kinase 5 D311N in Complex with CCG273441 | | Descriptor: | (3Z)-3-{[4-(2-chloroacetamido)-3,5-dimethyl-1H-pyrrol-2-yl]methylidene}-N-[(1R)-1-(4-fluorophenyl)ethyl]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, G protein-coupled receptor kinase 5 | | Authors: | Chen, Y, Tesmer, J.J.G. | | Deposit date: | 2023-09-21 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a new class of potent and highly selective G protein-coupled receptor kinase 5 inhibitors and structural insight from crystal structures of inhibitor complexes.
Eur.J.Med.Chem., 264, 2023
|
|
1YLJ
 
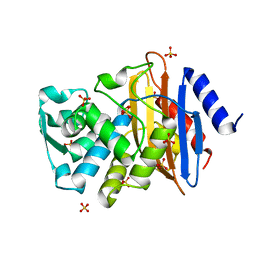 | | Atomic resolution structure of CTX-M-9 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YLY
 
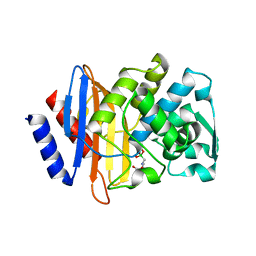 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with ceftazidime-like boronic acid | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase CTX-M-9 | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YLP
 
 | | Atomic resolution structure of CTX-M-27 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-27 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YLZ
 
 | | X-ray crystallographic structure of CTX-M-14 beta-lactamase complexed with ceftazidime-like boronic acid | | Descriptor: | BETA-LACTAMASE CTX-M-14, PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YMX
 
 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase covalently linked to cefoxitin | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, beta-lactamase CTX-M-9 | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-21 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YMS
 
 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with nafcinin-like boronic acid inhibitor | | Descriptor: | [(2-ETHOXY-1-NAPHTHOYL)AMINO]METHYLBORONIC ACID, beta-lactamase CTX-M-9 | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-21 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YLT
 
 | | Atomic resolution structure of CTX-M-14 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-14 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YM1
 
 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, PHOSPHATE ION, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YLW
 
 | | X-ray structure of CTX-M-16 beta-lactamase | | Descriptor: | CTX-M-16 beta-lactamase, SULFATE ION | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
3D6A
 
 | | Crystal structure of the 2H-phosphatase domain of Sts-2 in complex with tungstate. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Sts-2 protein, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of the 2H-phosphatase domain of Sts-2 reveals an acid-dependent phosphatase activity.
Biochemistry, 48, 2009
|
|
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK1
 
 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK3
 
 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK9
 
 | | High resolution structure of GCaMP3 dimer form 2 at pH 7.5 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, RCaMP, ... | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
