3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
4N4Z
 
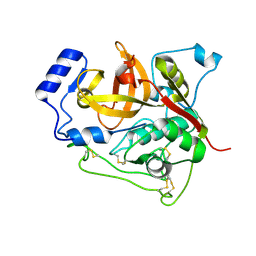 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
6RFU
 
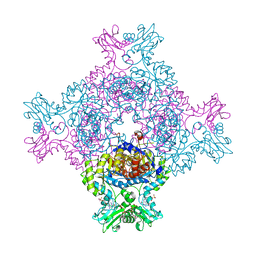 | | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of ATP and GMP as genuine co-factors | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase | | Authors: | Nass, K, Redecke, L, Perbandt, M, Yefanov, O, Gabdulkhakov, A, Duszenko, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors.
Nat Commun, 11, 2020
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | Descriptor: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z3U
 
 | | Crystal structure of SARS-CoV-2 Main Protease after incubation with Sulfo-Calpeptin | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, Calpeptin, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z2K
 
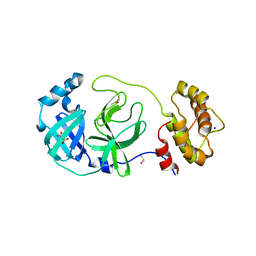 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections
To Be Published
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
8QKB
 
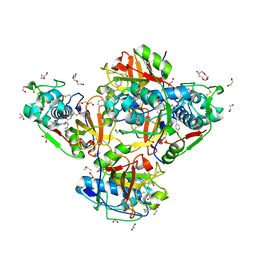 | | Crystal structure of human cathepsin L in complex with the vinyl sulfone inhibitor K777 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6FJS
 
 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5C6L
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using intense Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
5C6J
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
5C6I
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
5J7A
 
 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
8C77
 
 | | Human cathepsin L after reaction with the thiocarbazate inhibitor CID 16725315 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
5NJR
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 50s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
