1GO1
 
 | |
3H6P
 
 | | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis | | Descriptor: | ESAT-6 LIKE PROTEIN ESXS, ESAT-6-like protein esxR, GLYCEROL | | Authors: | Chan, S, Arbing, M, Phan, T, Kaufmann, M, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis
To be Published
|
|
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6U
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3JRN
 
 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
3K6X
 
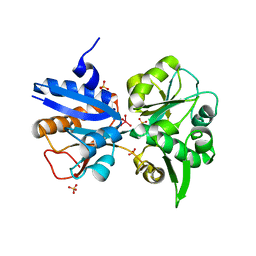 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6W
 
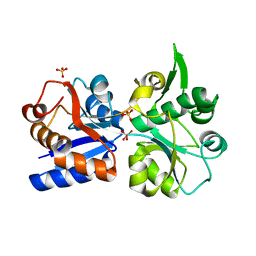 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1Z8F
 
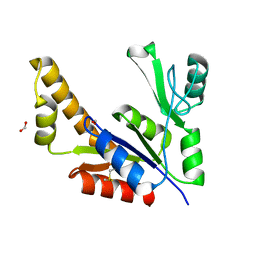 | | Guanylate Kinase Double Mutant A58C, T157C from Mycobacterium tuberculosis (Rv1389) | | Descriptor: | FORMIC ACID, Guanylate kinase | | Authors: | Chan, S, Sawaya, M.R, Choi, B, Zocchi, G, Perry, L.J, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis (Rv1389)
To be Published
|
|
1GO0
 
 | |
3H16
 
 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
6VGS
 
 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
1S4Q
 
 | | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis (Rv1389) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Guanylate kinase | | Authors: | Chan, S, Sawaya, M.R, Perry, L.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-01-16 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis
To be Published
|
|
3U2H
 
 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1Y67
 
 | |
3U2G
 
 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JRK
 
 | | NMR Structure and Epitope Mapping of Blo t 5 | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Chan, S, Mok, Y. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen
J.Immunol., 181, 2008
|
|
2A0A
 
 | |
5DLB
 
 | | Crystal structure of chaperone EspG3 of ESX-3 type VII secretion system from Mycobacterium marinum M | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Chan, S, Arbing, M.A, Kim, J, Kahng, S, Sawaya, M.R, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Variability of EspG Chaperones from Mycobacterial ESX-1, ESX-3, and ESX-5 Type VII Secretion Systems.
J. Mol. Biol., 431, 2019
|
|
4LF2
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with sulfate and magnesium | | Descriptor: | CARBONATE ION, MAGNESIUM ION, Ribulose bisphosphate carboxylase, ... | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
4LF1
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
2MOB
 
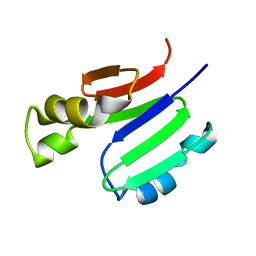 | | METHANE MONOOXYGENASE COMPONENT B | | Descriptor: | PROTEIN (METHANE MONOOXYGENASE REGULATORY PROTEIN B) | | Authors: | Chang, S.L, Wallar, B.J, Lipscomb, J.D, Mayo, K.H. | | Deposit date: | 1999-03-10 | | Release date: | 1999-08-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of component B from methane monooxygenase derived through heteronuclear NMR and molecular modeling.
Biochemistry, 38, 1999
|
|
6KAF
 
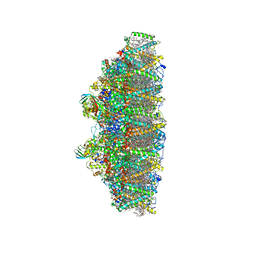 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
7ZLO
 
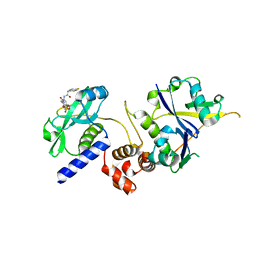 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 12 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
