7AS1
 
 | | Influenza A PB2 (F404Y mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, CHLORIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS2
 
 | | Influenza A PB2 (M431 mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS3
 
 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
3TNE
 
 | | The crystal structure of protease Sapp1p from Candida parapsilosis in complex with the HIV protease inhibitor ritonavir | | Descriptor: | RITONAVIR, Secreted aspartic protease | | Authors: | Dostal, J, Brynda, J, Hruskova-Heidingsfeldova, O, Pachl, P, Pichova, I, Rezacova, P. | | Deposit date: | 2011-09-01 | | Release date: | 2012-03-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of protease Sapp1p from Candida parapsilosis in complex with the HIV protease inhibitor ritonavir.
J Enzyme Inhib Med Chem, 27, 2012
|
|
3U7S
 
 | | HIV PR drug resistant patient's variant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, Pol polyprotein | | Authors: | Saskova, K.G, Rezacova, P, Konvalinka, J, Brynda, J, Kozisek, M. | | Deposit date: | 2011-10-14 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular characterization of clinical isolates of human immunodeficiency virus resistant to the protease inhibitor darunavir.
J.Virol., 83, 2009
|
|
4ZV5
 
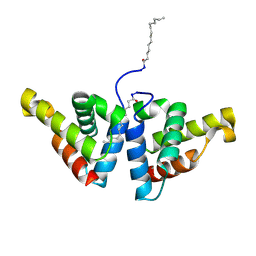 | | Crystal structure of N-myristoylated mouse mammary tumor virus matrix protein | | Descriptor: | MYRISTIC ACID, Matrix protein p10 | | Authors: | Zabransky, A, Dolezal, M, Dostal, J, Vanek, O, Hadravova, R, Stokrova, J, Brynda, J, Pichova, I. | | Deposit date: | 2015-05-18 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Myristoylation drives dimerization of matrix protein from mouse mammary tumor virus.
Retrovirology, 13, 2016
|
|
3ZHO
 
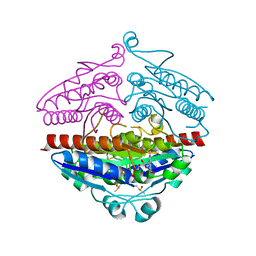 | | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOPROTEIN WRBA | | Authors: | Kishko, I, Lapkouski, M, Brynda, J, Kuty, M, Carey, J, Smatanova, I.K, Ettrich, R. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 A Resolution Crystal Structure of Escherichia Coli Wrba Holoprotein
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1JP5
 
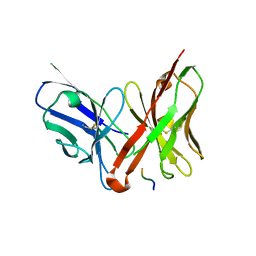 | | Crystal structure of the single-chain Fv fragment 1696 in complex with the epitope peptide corresponding to N-terminus of HIV-1 protease | | Descriptor: | epitope peptide corresponding to N-terminus of HIV-1 protease, single-chain Fv fragment 1696 | | Authors: | Rezacova, P, Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of HIV-1 and HIV-2 protease inhibition by a monoclonal antibody.
Structure, 9, 2001
|
|
6R5H
 
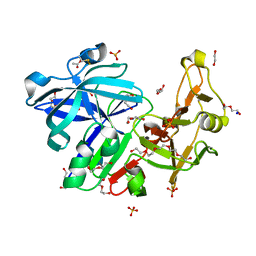 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
6R6A
 
 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endopeptidase, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
6S03
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | Descriptor: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
1N4X
 
 | | Structure of scFv 1696 at acidic pH | | Descriptor: | CHLORIDE ION, immunoglobulin heavy chain variable region, immunoglobulin kappa chain variable region | | Authors: | Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Rezacova, P, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2002-11-02 | | Release date: | 2003-06-10 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a single-chain Fv fragment of an antibody that inhibits the HIV-1 and HIV-2 proteases.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4O1G
 
 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
3QSD
 
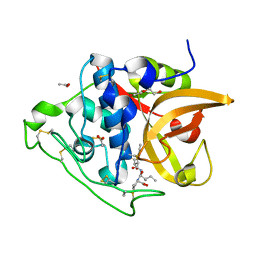 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with CA074 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
3S3Q
 
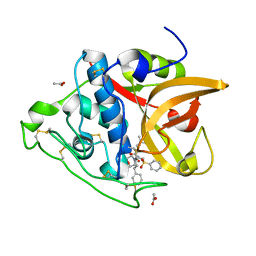 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11017 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
3S3R
 
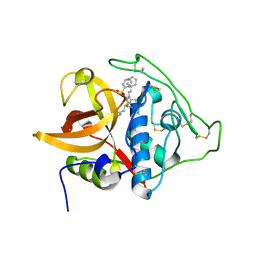 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11777 inhibitor | | Descriptor: | Cathepsin B-like peptidase (C01 family), Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
3TTP
 
 | | Structure of multiresistant HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Saskova, K.G, Brynda, J, Kozisek, M, Konvalinka, J, Rezacova, P. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Thermodynamic and structural analysis of HIV protease resistance to darunavir - analysis of heavily mutated patient-derived HIV-1 proteases.
Febs J., 281, 2014
|
|
7OYK
 
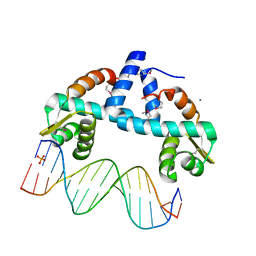 | | DNA-binding domain of CggR in complex with the DNA operator | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Central glycolytic genes regulator, ... | | Authors: | Novakova, M, Rezacova, P, Skerlova, J, Brynda, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-11-10 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural insight into DNA recognition by bacterial transcriptional regulators of the SorC/DeoR family.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7ZPY
 
 | |
7AGE
 
 | |
7AGB
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Candidapepsin, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7AGD
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB75 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7AGC
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB74 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
3L2C
 
 | | Crystal Structure of the DNA Binding Domain of FOXO4 Bound to DNA | | Descriptor: | FOXO consensus binding sequence, minus strand, plus strand, ... | | Authors: | Boura, E, Silhan, J, Sulc, M, Brynda, J, Obsilova, V, Obsil, T. | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA
Acta Crystallogr.,Sect.D, 66, 2010
|
|
