2JKD
 
 | | Structure of the yeast Pml1 splicing factor and its integration into the RES complex | | Descriptor: | GLYCEROL, PRE-MRNA LEAKAGE PROTEIN 1, SULFATE ION | | Authors: | Brooks, M.A, Dziembowski, A, Quevillon-Cheruel, S, Henriot, V, Faux, C, van Tilbeurgh, H, Seraphin, B. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Yeast Pml1 Splicing Factor and its Integration Into the Res Complex
Nucleic Acids Res., 37, 2009
|
|
4A8E
 
 | | The structure of a dimeric Xer recombinase from archaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROBABLE TYROSINE RECOMBINASE XERC-LIKE, ... | | Authors: | Brooks, M.A, ElArnaout, T, Duranda, D, Lisboa, J, Lazar, N, Raynal, B, vanTilbeurgh, H, Serre, M, Quevillon-Cheruel, S. | | Deposit date: | 2011-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Carboxy-Terminal Alpha N Helix of the Archaeal Xera Tyrosine Recombinase is a Molecular Switch to Control Site-Specific Recombination.
Plos One, 8, 2013
|
|
2W9J
 
 | | The crystal structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle | | Descriptor: | SIGNAL RECOGNITION PARTICLE SUBUNIT SRP14 | | Authors: | Brooks, M.A, Ravelli, R.B.G, McCarthy, A.A, Strub, K, Cusack, S. | | Deposit date: | 2009-01-25 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Srp14 from the Schizosaccharomyces Pombe Signal Recognition Particle.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VUG
 
 | | The structure of an archaeal homodimeric RNA ligase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PAB1020, ... | | Authors: | Brooks, M.A, Meslet-Cladiere, L, Graille, M, Kuhn, J, Blondeau, K, Myllykallio, H, van Tilbeurgh, H. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of an archaeal homodimeric ligase which has RNA circularization activity.
Protein Sci., 17, 2008
|
|
2XGL
 
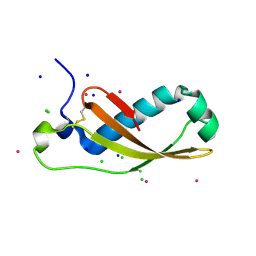 | | The X-ray structure of the Escherichia coli colicin M immunity protein demonstrates the presence of a disulphide bridge, which is functionally essential | | Descriptor: | CADMIUM ION, CHLORIDE ION, COLICIN-M IMMUNITY PROTEIN, ... | | Authors: | Gerard, F, Brooks, M.A, Barreteau, H, Touze, T, Graille, M, Bouhss, A, Blanot, D, Tilbeurgh, H.v, Mengin-Lecreulx, D. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-17 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure and Site-Directed Mutagenesis Analysis of the Escherichia Coli Colicin M Immunity Protein.
J.Bacteriol., 193, 2011
|
|
4UYK
 
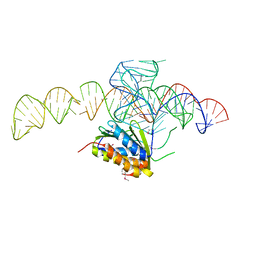 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
4UYJ
 
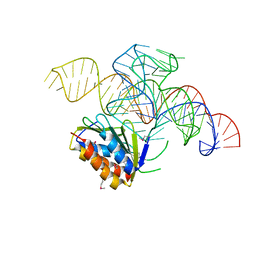 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
3UQZ
 
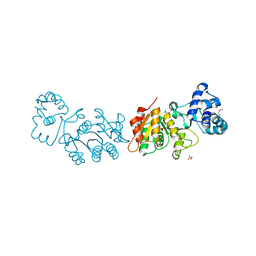 | |
