1F0N
 
 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85B | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1F0P
 
 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B WITH TREHALOSE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85-B, ... | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
1L9L
 
 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1FOU
 
 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S.N, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
1FOQ
 
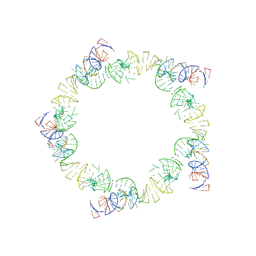 | | PENTAMERIC MODEL OF THE BACTERIOPHAGE PHI29 PROHEAD RNA | | Descriptor: | BACTERIOPHAGE PHI29 PROHEAD RNA | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | Authors: | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-26 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
1G9U
 
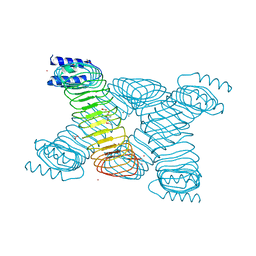 | | CRYSTAL STRUCTURE OF YOPM-LEUCINE RICH EFFECTOR PROTEIN FROM YERSINIA PESTIS | | Descriptor: | ACETATE ION, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Evdokimov, A.G, Anderson, D.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-11-28 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit.
J.Mol.Biol., 312, 2001
|
|
6V1S
 
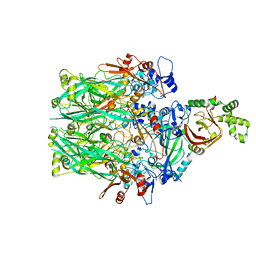 | | Structure of the Clostridioides difficile transferase toxin | | Descriptor: | ADP-ribosylating binary toxin enzymatic subunit CdtA, ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Sheedlo, M.J, Anderson, D.M, Thomas, A.K, Lacy, D.B. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural elucidation of theClostridioides difficiletransferase toxin reveals a single-site binding mode for the enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MRP
 
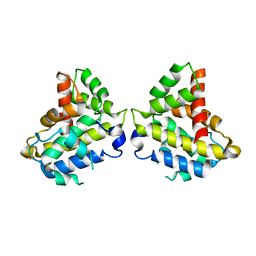 | |
6O2N
 
 | |
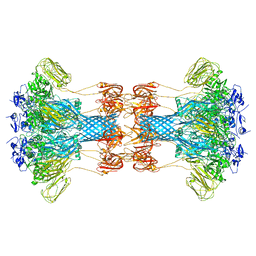
6O2M
 
 | |
6O2O
 
 | |
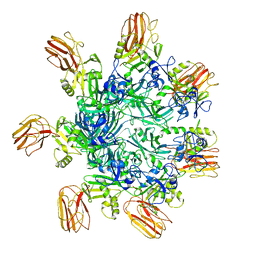
6OKU
 
 | |
6OKS
 
 | |
6OKT
 
 | |
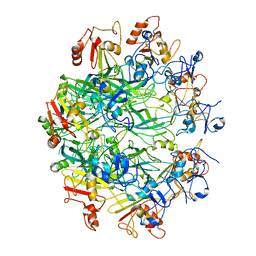
6OKR
 
 | |
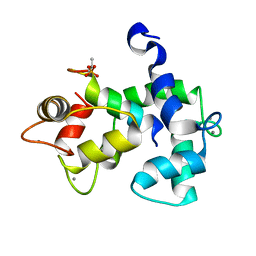
5J7J
 
 | |
1NOH
 
 | | The structure of bacteriophage phi29 scaffolding protein gp7 after prohead assembly | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.L, McMurray, C.T, L Anderson, D, Rossmann, M.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
4GID
 
 | | Structure of beta-secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
1XAE
 
 | | Crystal structure of wild type yellow fluorescent protein zFP538 from Zoanthus | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
1XA9
 
 | | Crystal structure of yellow fluorescent protein zFP538 K66M green mutant | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
6D85
 
 | |
