4WGK
 
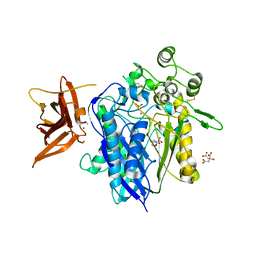 | | Crystal structure of human neutral ceramidase with Zn-bound phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Airola, M.V, Pulkoski-Gross, M.J, Obeid, L.M, Hannun, Y.A. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Structural Basis for Ceramide Recognition and Hydrolysis by Human Neutral Ceramidase.
Structure, 23, 2015
|
|
5UVG
 
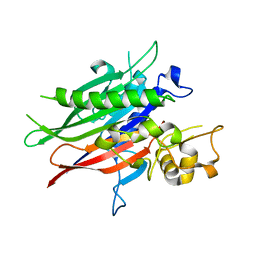 | | Crystal structure of the human neutral sphingomyelinase 2 (nSMase2) catalytic domain with insertion deleted and calcium bound | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase 3,Sphingomyelin phosphodiesterase 3 | | Authors: | Airola, M.V, Guja, K.E, Garcia-Diaz, M, Hannun, Y.A. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structure of human nSMase2 reveals an interdomain allosteric activation mechanism for ceramide generation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3LNR
 
 | |
4HI4
 
 | |
4I44
 
 | |
4I3M
 
 | | Aer2 poly-HAMP domains: L44H HAMP1 CW-lock mutant | | Descriptor: | Aerotaxis transducer Aer2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Airola, M.V, Sukomon, N, Crane, B.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HAMP Domain Conformers That Propagate Opposite Signals in Bacterial Chemoreceptors.
Plos Biol., 11, 2013
|
|
8EXD
 
 | |
6TZZ
 
 | |
6U8Z
 
 | |
6TZY
 
 | |
7KIL
 
 | |
7KIH
 
 | |
7KIQ
 
 | |
7LHK
 
 | |
7LPO
 
 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytoplasmic protein, MAGNESIUM ION | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LPQ
 
 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 9 | | Descriptor: | Cytoplasmic protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LPP
 
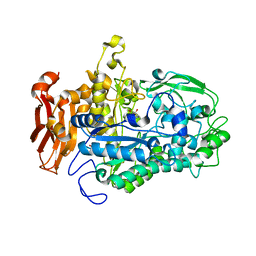 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 1 | | Descriptor: | 4-(hydroxymethyl)-1-[[2-(3-methoxyphenyl)-1,3-thiazol-5-yl]methyl]piperidin-4-ol, Cytoplasmic protein, GLYCEROL, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
