5EJ1
 
 | | Pre-translocation state of bacterial cellulose synthase | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Moragn, J.L.W, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP6
 
 | | Chlorella virus hyaluronan synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SPA
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP9
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
6TZK
 
 | | Bacterial cellulose synthase outermembrane channel BcsC with terminal TPR repeat | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Acheson, J.F, Derewenda, Z, Zimmer, J. | | Deposit date: | 2019-08-12 | | Release date: | 2019-10-23 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Architecture of the Cellulose Synthase Outer Membrane Channel and Its Association with the Periplasmic TPR Domain.
Structure, 27, 2019
|
|
6OIH
 
 | |
8DNC
 
 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen and ADP | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DKU
 
 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, Transport permease protein | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DKY
 
 | |
8DN8
 
 | |
8DL0
 
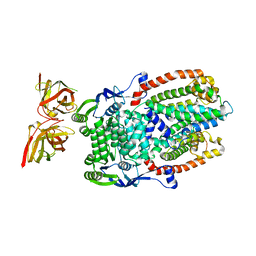 | |
8DNE
 
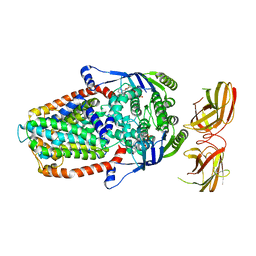 | |
8DOU
 
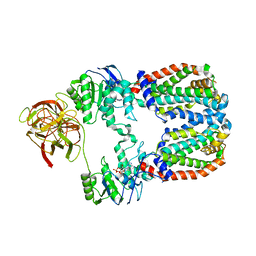 | |
1P7B
 
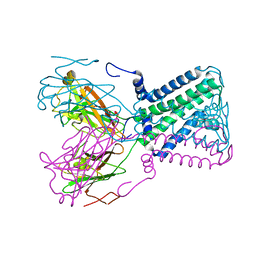 | | Crystal structure of an inward rectifier potassium channel | | Descriptor: | POTASSIUM ION, integral membrane channel and cytosolic domains | | Authors: | Kuo, A, Gulbis, J.M, Antcliff, J.F, Rahman, T, Lowe, E.D, Zimmer, J, Cuthbertson, J, Ashcroft, F.M, Ezaki, T, Doyle, D.A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the potassium channel KirBac1.1 in the closed state.
Science, 300, 2003
|
|
8DQK
 
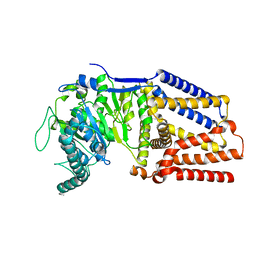 | | Intermediate resolution structure of barley (1,3;1,4)-beta-glucan synthase CslF6. | | Descriptor: | Cellulose synthase-like CslF6 | | Authors: | Ho, R, Purushotham, P, Zimmer, J. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of mixed-linkage glucan biosynthesis by barley cellulose synthase-like CslF6 (1,3;1,4)-beta-glucan synthase.
Sci Adv, 8, 2022
|
|
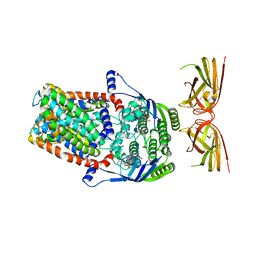
7K2T
 
 | |
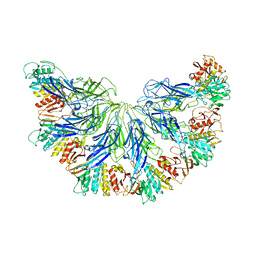
7L2Z
 
 | |
4P02
 
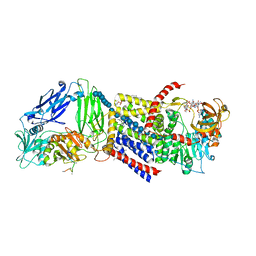 | | Structure of Bacterial Cellulose Synthase with cyclic-di-GMP bound. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cellulose Synthase subunit A, ... | | Authors: | Morgan, J.L.W, McNamara, J.T, Zimmer, J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4P00
 
 | | Bacterial Cellulose Synthase in complex with cyclic-di-GMP and UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cellulose Synthase A subunit, ... | | Authors: | Morgan, J.L.W, McNamara, J.T, Zimmer, J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7PCD
 
 | | HER2 IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 1-[4-[4-[[3,5-bis(chloranyl)-4-([1,2,4]triazolo[1,5-a]pyridin-7-yloxy)phenyl]amino]pyrimido[5,4-d]pyrimidin-6-yl]piperazin-1-yl]-4-(3-fluoranylazetidin-1-yl)butan-1-one, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Bader, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of potent and selective HER2 inhibitors with efficacy against HER2 exon 20 insertion-driven tumors, which preserve wild-type EGFR signaling.
Nat Cancer, 3, 2022
|
|
2WLL
 
 | | POTASSIUM CHANNEL FROM BURKHOLDERIA PSEUDOMALLEI | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, MAGNESIUM ION, POTASSIUM CHANNEL, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, VandenBerg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
4QTB
 
 | | Structure of human ERK1 in complex with SCH772984 revealing a novel inhibitor-induced binding pocket | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
4QTE
 
 | | Structure of ERK2 in complex with VTX-11e, 4-{2-[(2-CHLORO-4-FLUOROPHENYL)AMINO]-5-METHYLPYRIMIDIN-4-YL}-N-[(1S)-1-(3-CHLOROPHENYL)-2-HYDROXYETHYL]-1H-PYRROLE-2-CARBOXAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[(2-chloro-4-fluorophenyl)amino]-5-methylpyrimidin-4-yl}-N-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
