4ZIC
 
 | |
1C8M
 
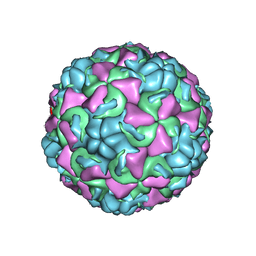 | | REFINED CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS 16 COMPLEXED WITH VP63843 (PLECONARIL), AN ANTI-PICORNAVIRAL DRUG CURRENTLY IN CLINICAL TRIALS | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, HUMAN RHINOVIRUS 16 COAT PROTEIN, ZINC ION | | Authors: | Chakravarty, S, Bator, C.M, Pevear, D.C, Diana, G.D, Rossmann, M.G. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE REFINED STRUCTURE OF A PICORNAVIRUS INHIBITOR CURRENTLY IN CLINICAL TRIALS, WHEN COMPLEXED WITH HUMAN RHINOVIRUS 16
to be published
|
|
4GOS
 
 | | Crystal structure of human B7-H4 IgV-like domain | | Descriptor: | V-set domain-containing T-cell activation inhibitor 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vigdorovich, V, Ramagopal, U, Bhosle, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and cancer immunotherapy of the B7 family member B7x.
Cell Rep, 9, 2014
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1QJU
 
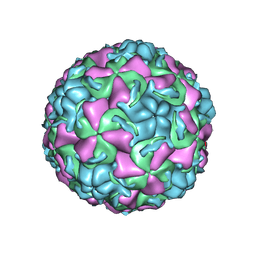 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJY
 
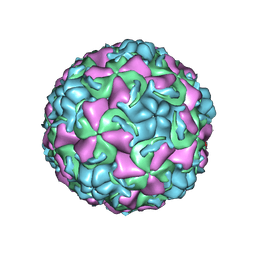 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJX
 
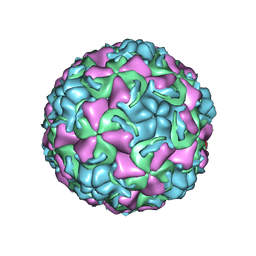 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3LD8
 
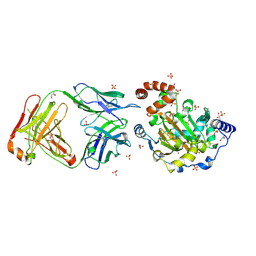 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LDB
 
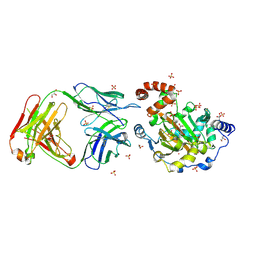 | | Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8IKL
 
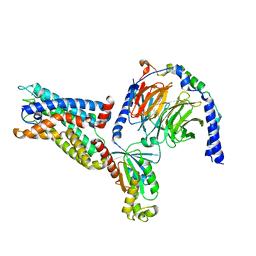 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
6NCU
 
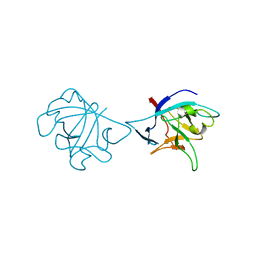 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8IX0
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX1
 
 | | Cryo-EM structure of protonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX2
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWY
 
 | | Cryo-EM structure of protonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWZ
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWX
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
2M01
 
 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | Descriptor: | Protease inhibitor LmKTT-1a | | Authors: | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
7EQB
 
 | |
