5XJF
 
 | | Crystal structure of fucosylated IgG Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5XFW
 
 | | Crystal structures of FMN-free form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
5WV4
 
 | |
2D49
 
 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | Descriptor: | chitinase C | | Authors: | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | Deposit date: | 2005-10-11 | | Release date: | 2006-10-11 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
1HS7
 
 | | VAM3P N-TERMINAL DOMAIN SOLUTION STRUCTURE | | Descriptor: | SYNTAXIN VAM3 | | Authors: | Dulubova, I, Yamaguchi, T, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2000-12-24 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Vam3p structure reveals conserved and divergent properties of syntaxins.
Nat.Struct.Biol., 8, 2001
|
|
3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4I
 
 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4J
 
 | | Crystal Structure of human DAAO in complex with coumpound 12 | | Descriptor: | 3-hydroxy-5-(2-phenylethyl)pyridin-2(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3WMG
 
 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Sakiyama, K, Hipolito, C.J, Fujioka, A, Hirokane, R, Ikeguchi, K, Watanabe, B, Hirtake, J, Kimura, Y, Suga, H, Ueda, K, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WME
 
 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMF
 
 | | Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WNX
 
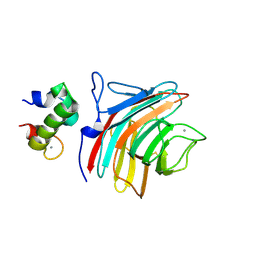 | | Crystal structure of ERGIC-53/MCFD2, Calcium/Man3-bound form | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53, ... | | Authors: | Satoh, T, Suzuki, K, Yamaguchi, T, Kato, K. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Disparate Sugar-Binding Specificities in the Homologous Cargo Receptors ERGIC-53 and VIP36
Plos One, 9, 2014
|
|
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3WK5
 
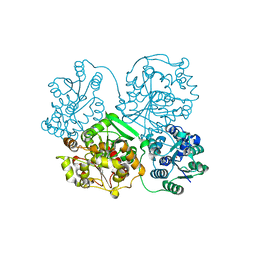 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 2-cyclopentyl-N-(1,3-thiazol-2-yl)acetamide, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WKD
 
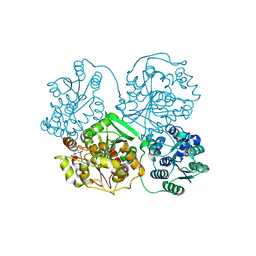 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-[2-(morpholin-4-yl)phenyl]thiophene-3-carboxamide, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WKC
 
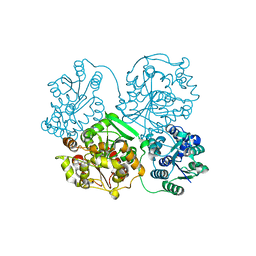 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 4-{2,5-dimethyl-1-[(2R)-tetrahydrofuran-2-ylmethyl]-1H-pyrrol-3-yl}-1,3-thiazol-2-amine, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WKE
 
 | | Crystal structure of soluble epoxide hydrolase in complex with t-AUCB | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WK8
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 6-(trifluoromethyl)-1,3-benzothiazol-2-amine, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WK9
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 5-(4-bromobenzyl)-1,3-thiazol-2-amine, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WK4
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 1-[(1R)-1-cyclopropylethyl]-3-phenylurea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WK7
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 2-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazole, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WKA
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 6-amino-1-methyl-5-(piperidin-1-yl)pyrimidine-2,4(1H,3H)-dione, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WKB
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | 4-benzyl-3,4-dihydroquinoxalin-2(1H)-one, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-18 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
3WK6
 
 | | Crystal structure of soluble epoxide hydrolase in complex with fragment inhibitor | | Descriptor: | (5R)-5-methyl-N-(2-phenylethyl)-4,5-dihydro-1,3-thiazol-2-amine, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Amano, Y, Yamaguchi, T, Tanabe, E. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into binding of inhibitors to soluble epoxide hydrolase gained by fragment screening and X-ray crystallography.
Bioorg.Med.Chem., 22, 2014
|
|
6A22
 
 | | Ternary complex of Human ROR gamma Ligand Binding Domain With Compound T. | | Descriptor: | 2-[2-[1-~{tert}-butyl-5-(4-methoxyphenyl)pyrazol-4-yl]-1,3-thiazol-4-yl]-~{N}-(oxan-4-ylmethyl)ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ternary crystal structure of human ROR gamma ligand-binding-domain, an inhibitor and corepressor peptide provides a new insight into corepressor interaction
Sci Rep, 8, 2018
|
|
