4QC6
 
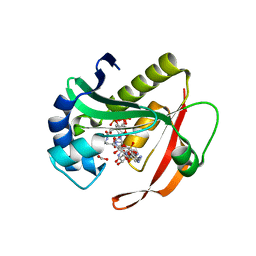 | | Crystal structure of aminoglycoside 6'-acetyltransferase-Ie | | Descriptor: | (3R,5S,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3,5-diphosphaheptadecane-17-sulfinic acid 3,5-dioxide (non-preferred name), Bifunctional AAC/APH, FORMIC ACID, ... | | Authors: | Smith, C.A, Toth, M, Weiss, T.M, Frase, H, Vakulenko, S.B. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the bifunctional aminoglycoside-resistance enzyme AAC(6')-Ie-APH(2'')-Ia revealed by crystallographic and small-angle X-ray scattering analysis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5DZ2
 
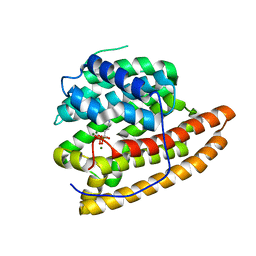 | | Geosmin synthase from Streptomyces coelicolor N-terminal domain complexed with three Mg2+ ions and alendronic acid | | Descriptor: | ALENDRONATE, Germacradienol/geosmin synthase, MAGNESIUM ION | | Authors: | Harris, G.G, Lombardi, P.M, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K.W, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
5DW7
 
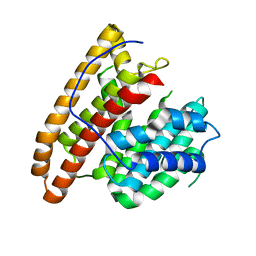 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
4JEU
 
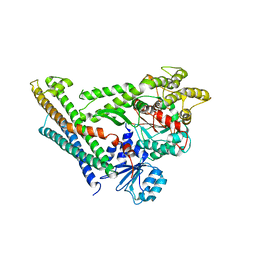 | | Crystal Structure of Munc18a and Syntaxin1 with native N-terminus complex | | Descriptor: | Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Colbert, K.N, Hattendorf, D.A, Weiss, T.M, Burkhardt, P, Fasshauer, D, Weis, W.I. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JEH
 
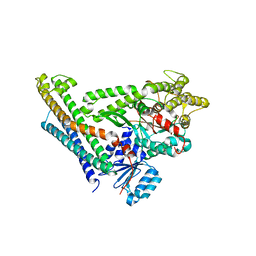 | | Crystal Structure of Munc18a and Syntaxin1 lacking N-peptide complex | | Descriptor: | Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Colbert, K.N, Hattendorf, D.A, Weiss, T.M, Burkhardt, P, Fasshauer, D, Weis, W.I. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6DNK
 
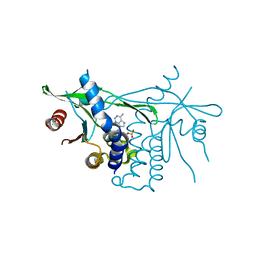 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
4XKH
 
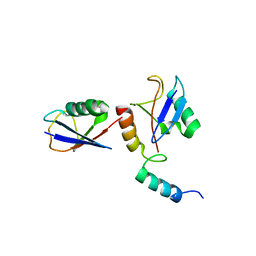 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | Descriptor: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | Authors: | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | Deposit date: | 2015-01-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
4IFQ
 
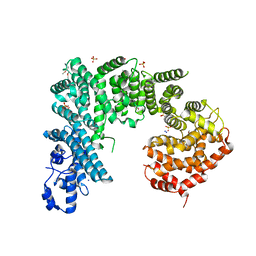 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
6CFF
 
 | | Stimulator of Interferon Genes Human | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
6CY7
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
4NUZ
 
 | | Crystal structure of a glycosynthase mutant (D233Q) of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NUY
 
 | | Crystal structure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6P5T
 
 | | Surface-layer (S-layer) RsaA protein from Caulobacter crescentus bound to strontium and iodide | | Descriptor: | IODIDE ION, S-layer protein, STRONTIUM ION | | Authors: | Chan, A.C, Herrmann, J, Smit, J, Wakatsuki, S, Murphy, M.E. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bacterial surface layer protein exploits multistep crystallization for rapid self-assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DZZ
 
 | |
7T2Y
 
 | | X-ray structure of a designed cold unfolding four helix bundle | | Descriptor: | Designed cold unfolding four helix bundle | | Authors: | Harrison, J.S, Kuhlman, B, Szyperski, T, Premkumar, L, Maguire, J, Pulavarti, S, Yuen, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
2KLL
 
 | | Solution structure of human interleukin-33 | | Descriptor: | Interleukin-33 | | Authors: | Lingel, A, Fairbrother, W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes.
Structure, 17, 2009
|
|
