6EXE
 
 | |
6EXC
 
 | |
6EXA
 
 | |
6EXD
 
 | |
6EXB
 
 | |
6GEB
 
 | |
2GR8
 
 | | Hia 1022-1098 | | Descriptor: | Adhesin | | Authors: | Meng, G, Waksman, G. | | Deposit date: | 2006-04-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter.
Embo J., 25, 2006
|
|
2GR7
 
 | | Hia 992-1098 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Adhesin | | Authors: | Meng, G, Waksman, G. | | Deposit date: | 2006-04-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter.
Embo J., 25, 2006
|
|
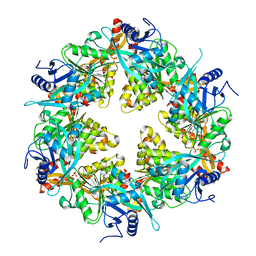
2BHM
 
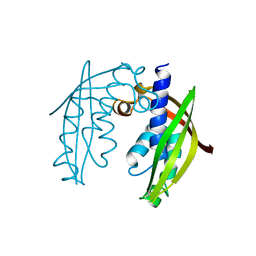 | | Crystal structure of VirB8 from Brucella suis | | Descriptor: | TYPE IV SECRETION SYSTEM PROTEIN VIRB8 | | Authors: | Bayliss, R, Baron, C, Waksman, G. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2PT7
 
 | | Crystal structure of Cag VirB11 (HP0525) and an inhibitory protein (HP1451) | | Descriptor: | Cag-alfa, Hypothetical protein | | Authors: | Hare, S, Fischer, W, Williams, R, Terradot, L, Bayliss, R, Haas, R, Waksman, G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase.
Embo J., 26, 2007
|
|
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2UY7
 
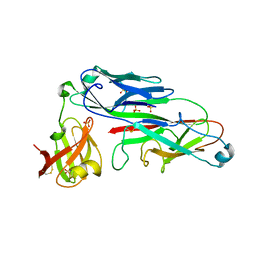 | | Crystal structure of the P pilus rod subunit PapA | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN, PERIPLASMID CHAPERONE PAPD PROTEIN, SULFATE ION | | Authors: | Verger, D, Bullitt, E, Hultgren, S.J, Waksman, G. | | Deposit date: | 2007-04-02 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the P Pilus Rod Subunit Papa.
Plos Pathog., 3, 2007
|
|
2UY6
 
 | | Crystal structure of the P pilus rod subunit PapA | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN, PERIPLASMID CHAPERONE PAPD PROTEIN | | Authors: | Verger, D, Bullitt, E, Hultgren, S.J, Waksman, G. | | Deposit date: | 2007-04-02 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the P Pilus Rod Subunit Papa.
Plos Pathog., 3, 2007
|
|
1IS0
 
 | | Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor | | Descriptor: | AY0 GLU GLU ILE peptide, Tyrosine-protein kinase transforming protein SRC | | Authors: | Davidson, J.P, Lubman, O, Rose, T, Waksman, G, Martin, S.F. | | Deposit date: | 2001-11-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calorimetric and structural studies of 1,2,3-trisubstituted cyclopropanes as conformationally constrained peptide inhibitors of Src SH2 domain binding.
J.Am.Chem.Soc., 124, 2002
|
|
2VQI
 
 | | Structure of the P pilus usher (PapC) translocation pore | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE USHER PROTEIN PAPC | | Authors: | Remaut, H, Tang, C, Henderson, N.S, Pinkner, J.S, Wang, T, Hultgren, S.J, Thanassi, D.G, Li, H, Waksman, G. | | Deposit date: | 2008-03-16 | | Release date: | 2008-05-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Fiber formation across the bacterial outer membrane by the chaperone/usher pathway.
Cell, 133, 2008
|
|
5KTQ
 
 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
5LER
 
 | | Structure of the bacterial sex F pilus (13.2 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5LEG
 
 | | Structure of the bacterial sex F pilus (pED208) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5LFB
 
 | | Structure of the bacterial sex F pilus (12.5 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
1KA8
 
 | | Crystal Structure of the Phage P4 Origin-Binding Domain | | Descriptor: | putative P4-specific DNA primase | | Authors: | Yeo, H.J, Ziegelin, G, Korolev, S, Calendar, R, Lanka, E, Waksman, G. | | Deposit date: | 2001-10-31 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins.
Mol.Microbiol., 43, 2002
|
|
7OVB
 
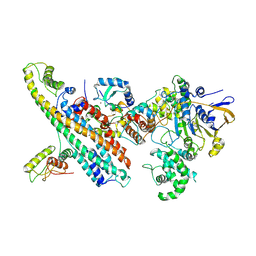 | | L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains | | Descriptor: | DotY, DotZ, IcmJ (DotN), ... | | Authors: | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proteins DotY and DotZ modulate the dynamics and localization of the type IVB coupling complex of Legionella pneumophila.
Mol.Microbiol., 117, 2022
|
|
2J7L
 
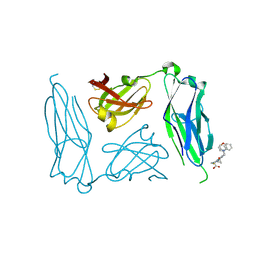 | | E. coli P Pilus chaperone PapD in complex with a pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD | | Authors: | Remaut, H, Pinkner, J.S, Hultgren, S.J, Almqvist, F, Waksman, G. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationally Designed Small Compounds Inhibit Pilus Biogenesis in Uropathogenic Bacteria.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2J2Z
 
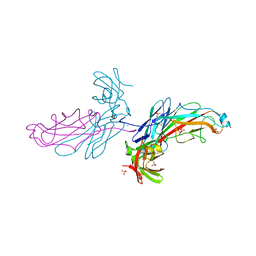 | | X-Ray Structure of the Chaperone PapD in complex with the Pilus terminator subunit PapH at 2.3 Angstrom resolution | | Descriptor: | CHAPERONE PROTEIN PAPD, COBALT (II) ION, PAP FIMBRIAL MINOR PILIN PROTEIN, ... | | Authors: | Verger, D, Miller, E, Remaut, H, Waksman, G, Hultgren, S. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of P Pilus Termination in Uropathogenic Escherichia Coli.
Embo Rep., 7, 2006
|
|
2KT6
 
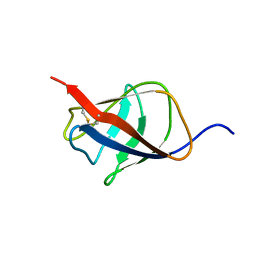 | | Structural homology between the C-terminal domain of the PapC usher and its plug | | Descriptor: | Outer membrane usher protein papC | | Authors: | Ford, B, Rego, A, Ragan, T.J, Pinkner, J, Dodson, K, Driscoll, P.C, Hultgren, S, Waksman, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Homology between the C-Terminal Domain of the PapC Usher and Its Plug.
J.Bacteriol., 192, 2010
|
|
2KTQ
 
 | | OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-07-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
