6XAH
 
 | | Structure of a Stable Interstrand DNA Crosslink Involving an dA Amino Group and an Abasic Site | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*GP*AP*AP*CP*(AAB)P*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*TP*AP*AP*GP*TP*TP*CP*AP*TP*CP*TP*A)-3') | | Authors: | Kellum Jr, A.H, Qiu, D, Voehler, M.W, Martin, W.J, Gates, K.S, Stone, M.P. | | Deposit date: | 2020-06-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable Interstrand DNA Cross-Link Involving a beta- N -Glycosyl Linkage Between an N 6 -dA Amino Group and an Abasic Site.
Biochemistry, 60, 2021
|
|
2LIA
 
 | | Solution NMR structure of a DNA dodecamer containing the 7-aminomethyl-7-deaza-2'-deoxyguanosine adduct | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*(2LA)P*CP*GP*CP*TP*CP*TP*C)-3') | | Authors: | Szulik, M.W, Ganguly, M, Wang, R, Gold, B, Stone, M.P. | | Deposit date: | 2011-08-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Site-Specific Stabilization of DNA by a Tethered Major Groove Amine, 7-Aminomethyl-7-deaza-2'-deoxyguanosine.
Biochemistry, 52, 2013
|
|
5KZO
 
 | |
8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
2KH3
 
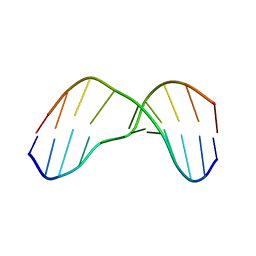 | | NMR Structure of Aflatoxin Formamidopyrimidine alpha-anomer in duplex DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3', 5'-D(*TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural perturbations induced by the alpha-anomer of the aflatoxin B(1) formamidopyrimidine adduct in duplex and single-strand DNA
J.Am.Chem.Soc., 131, 2009
|
|
2KH4
 
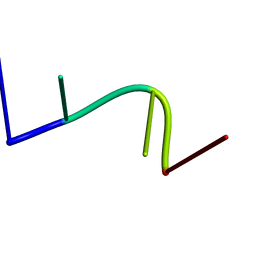 | | Aflatoxin Formamidopyrimidine alpha anomer in single strand DNA | | Descriptor: | 5'-D(*CP*TP*(FAG)P*A)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural perturbations induced by the alpha-anomer of the aflatoxin B(1) formamidopyrimidine adduct in duplex and single-strand DNA
J.Am.Chem.Soc., 131, 2009
|
|
