8TCU
 
 | |
7MYB
 
 | |
7MY9
 
 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Campbell, A.C. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MWT
 
 | |
7MWU
 
 | |
7MWV
 
 | |
7NA0
 
 | |
4Q72
 
 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D779Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Pemberton, T.A, Luo, M. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
4Q73
 
 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D778Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
5CDH
 
 | |
5CKU
 
 | |
4O8A
 
 | |
2RBF
 
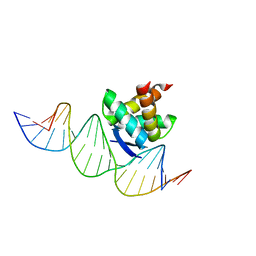 | | Structure of the ribbon-helix-helix domain of Escherichia coli PutA (PutA52) complexed with operator DNA (O2) | | Descriptor: | Bifunctional protein putA, DNA (5'-D(*DTP*DT*DTP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DCP*DAP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DGP*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DAP*DAP*DA)-3') | | Authors: | Tanner, J.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
J.Mol.Biol., 381, 2008
|
|
6MVR
 
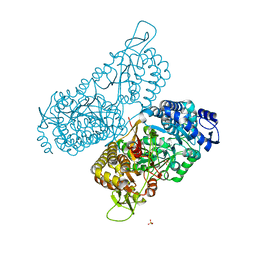 | | Structure of a bacterial ALDH16 | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6O4E
 
 | | Structure of ALDH7A1 mutant N167S complexed with NAD | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4F
 
 | | Structure of ALDH7A1 mutant N167S complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4H
 
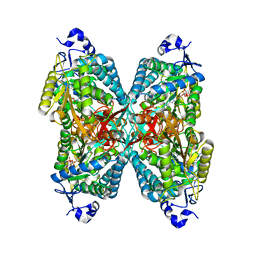 | | Structure of ALDH7A1 mutant A171V complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
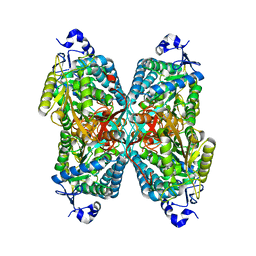
6O4G
 
 | |
6O4C
 
 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4K
 
 | | Structure of ALDH7A1 mutant E399Q complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4D
 
 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4B
 
 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4L
 
 | | Structure of ALDH7A1 mutant E399D complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
4GDE
 
 | |
