3EZJ
 
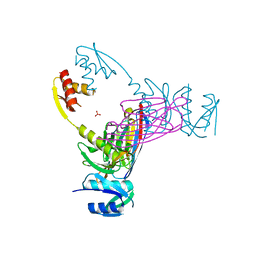 | | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody | | Descriptor: | CHLORIDE ION, General secretion pathway protein GspD, NANOBODY NBGSPD_7, ... | | Authors: | Korotkov, K.V, Pardon, E, Steyaert, J, Hol, W.G. | | Deposit date: | 2008-10-22 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Structure, 17, 2009
|
|
5M95
 
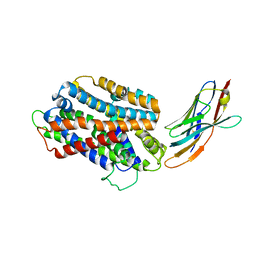 | | STAPHYLOCOCCUS CAPITIS DIVALENT METAL ION TRANSPORTER (DMT) IN COMPLEX WITH MANGANESE | | Descriptor: | CAMELID ANTIBODY FRAGMENT, NANOBODY, Divalent metal cation transporter MntH, ... | | Authors: | Ehrnstorfer, I.A, Geertsma, E.R, Pardon, E, Steyaert, J, Dutzler, R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-30 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure Of A Slc11 (Nramp) Transporter Reveals The Basis For Transition-Metal Ion Transport.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5NBD
 
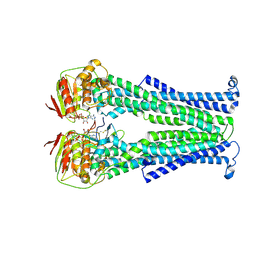 | | PglK flippase in complex with inhibitory nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | Authors: | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
3FZ0
 
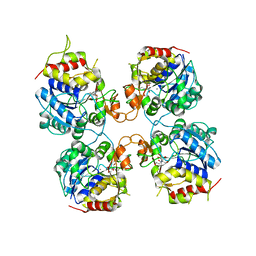 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | Deposit date: | 2009-01-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
4BIR
 
 | | RIBONUCLEASE T1: FREE HIS92GLN MUTANT | | Descriptor: | CALCIUM ION, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | Doumen, J, Steyaert, J. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
4MQS
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, Nanobody 9-8 | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
6GCI
 
 | | Structure of the bongkrekic acid-inhibited mitochondrial ADP/ATP carrier | | Descriptor: | Bongkrekic acid, CARDIOLIPIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Ruprecht, J.J, King, M.S, Pardon, E, Aleksandrova, A.A, Zogg, T, Crichton, P.G, Steyaert, J, Kunji, E.R.S. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Cell, 176, 2019
|
|
4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
6GJQ
 
 | | human NBD1 of CFTR in complex with nanobody T27 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, Nanobody T27 | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GKD
 
 | | human NBD1 of CFTR in complex with nanobodies D12 and G3a | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
5JQH
 
 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
4HOH
 
 | | RIBONUCLEASE T1 (THR93ALA MUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
4CND
 
 | | Crystal structure of E.coli TrmJ | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (CYTIDINE/URIDINE-2'-O-)-METHYLTRANSFERASE TRMJ | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4CNE
 
 | | Crystal structure of E.coli TrmJ in complex with S-adenosyl-L- homocysteine | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, TRNA (CYTIDINE/URIDINE-2'-O-)-METHYLTRANSFERASE TRMJ | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4CNG
 
 | | Crystal structure of Sulfolobus acidocaldarius TrmJ in complex with S-adenosyl-L-Homocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SPOU RRNA METHYLASE | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4CNF
 
 | | Crystal structure of Sulfolobus acidocaldarius TrmJ | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
4N9O
 
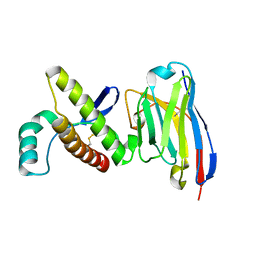 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
5OJM
 
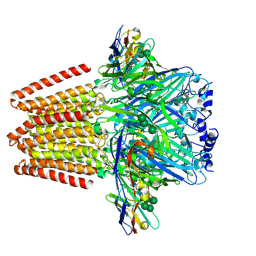 | | Structure of a chimaeric beta3-alpha5 GABAA receptor in complex with nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human GABAA receptor chimera beta3-alpha5,Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit alpha-5, Nanobody Nb25, ... | | Authors: | Miller, P.S, Scott, S, Masiulis, S, De Colibus, L, Pardon, E, Steyaert, J, Aricescu, A.R. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for GABAA receptor potentiation by neurosteroids.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O8F
 
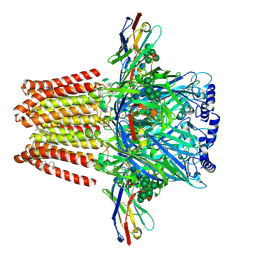 | | Structure of a chimaeric beta3-alpha5 GABAA receptor in complex with nanobody Nb25 and pregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, Nanobody Nb25, ... | | Authors: | Miller, P.S, Scott, S, Masiulis, S, De Colibus, L, Pardon, E, Steyaert, J, Aricescu, A.R. | | Deposit date: | 2017-06-13 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for GABAA receptor potentiation by neurosteroids.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1HZ1
 
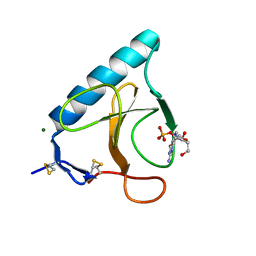 | | RIBONUCLEASE T1 V16A MUTANT IN COMPLEX WITH MG2+ | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, MAGNESIUM ION, RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1I0V
 
 | | Ribonuclease T1 in complex with 2'GMP (form I crystal) | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.234 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1I0X
 
 | | RIBONUCLEASE T1 IN COMPLEX WITH 2'GMP (FORM II CRYSTAL) | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
