7FJD
 
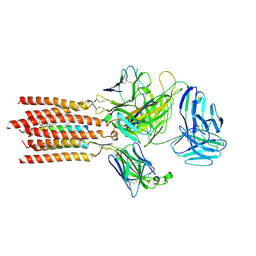 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7WRZ
 
 | | Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5840H, BD55-5840L, ... | | Authors: | Zhang, Z.Z, Xiao, J.J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7WRO
 
 | | Local structure of BD55-3372 and delta spike | | Descriptor: | 3372H, 3372L, Spike protein S1 | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7WRL
 
 | |
7WR8
 
 | |
6KPC
 
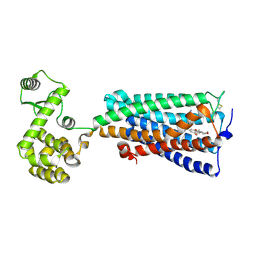 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPF
 
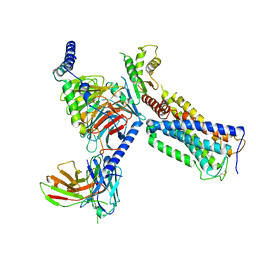 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPG
 
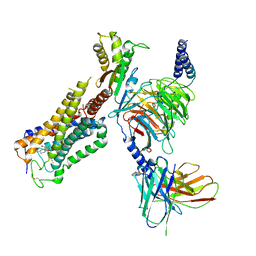 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
2LHV
 
 | | Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2012-06-27 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LHY
 
 | | Di-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LI1
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LHX
 
 | | Di-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
3OWD
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone II | | Descriptor: | Heat shock protein HSP 90-alpha, N-{[1-(5-chloro-2,4-dihydroxyphenyl)-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]methyl}naphthalene-1-sulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OWB
 
 | | Crystal Structure of HSP90 with VER-49009 | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW6
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone I | | Descriptor: | 1-(2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2LI2
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LHW
 
 | | Tri-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LI0
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LHZ
 
 | | Di-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2MMP
 
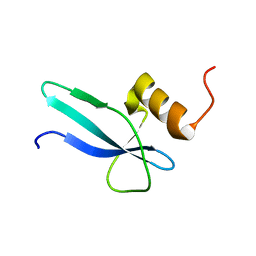 | | Solution structure of a ribosomal protein | | Descriptor: | Uncharacterized protein | | Authors: | Feng, Y. | | Deposit date: | 2014-03-17 | | Release date: | 2014-06-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure determination of archaea-specific ribosomal protein L46a reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 450, 2014
|
|
2Q8O
 
 | |
4EF5
 
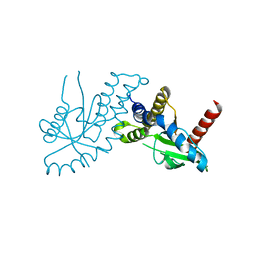 | | Crystal structure of STING CTD | | Descriptor: | Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
4HR9
 
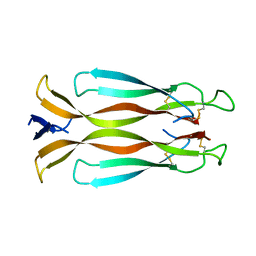 | | Human interleukin 17A | | Descriptor: | Interleukin-17A | | Authors: | Liu, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-05-22 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
4HSA
 
 | | Structure of interleukin 17a in complex with il17ra receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Liu, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
7DTZ
 
 | | FGFR4 complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | Authors: | Chen, X.J, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
