4ORE
 
 | |
6BO8
 
 | | Cryo-EM structure of human TRPV6 in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BOB
 
 | | Cryo-EM structure of rat TRPV6* in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BO9
 
 | | Cryo-EM structure of human TRPV6 in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BOA
 
 | | Cryo-EM structure of human TRPV6-R470E in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
7ESS
 
 | | Structure-guided studies of the Holliday junction resolvase RuvX provide novel insights into ATP-stimulated cleavage of branched DNA and RNA substrates | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Thakur, M, Mohan, D, Singh, A.K, Agarwal, A, Gopal, B, Muniyappa, K. | | Deposit date: | 2021-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Novel insights into ATP-Stimulated Cleavage of branched DNA and RNA Substrates through Structure-Guided Studies of the Holliday Junction Resolvase RuvX.
J.Mol.Biol., 433, 2021
|
|
7EWO
 
 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
7K4D
 
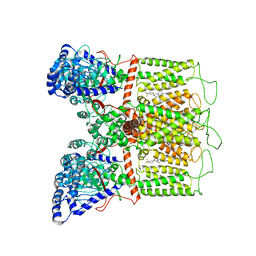 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 3OG | | Descriptor: | 5-[(4-{trans-4-hydroxy-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4F
 
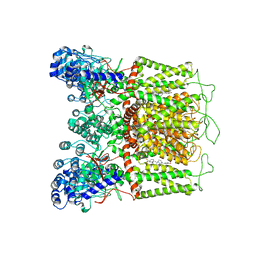 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 31 | | Descriptor: | 5-[(4-{cis-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4B
 
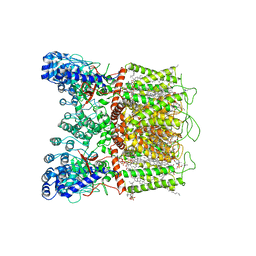 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor cis-22a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[cis-4-(3-methylphenyl)cyclohexyl]-4-(pyridin-3-yl)piperazine, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4C
 
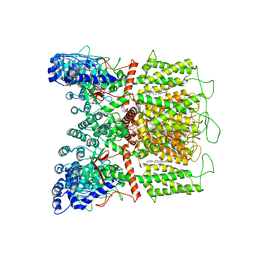 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromopyridin-3-yl)-4-[cis-4-(3-methylphenyl)cyclohexyl]piperazine, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4E
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 30 | | Descriptor: | 5-({4-[(1R,4S)-3'-methyl[1,2,3,4-tetrahydro[1,1'-biphenyl]]-4-yl]piperazin-1-yl}methyl)pyridin-2(1H)-one, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4A
 
 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
2QPK
 
 | | Crystal structure of the complex of bovine lactoperoxidase with salicylhydroxamic acid at 2.34 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-07-24 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
4Z1T
 
 | |
4Z15
 
 | | MIF in complex with 3-(2-furylmethyl)-2-thioxo-1,3-thiazolan-4-one | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Cho, T.Y. | | Deposit date: | 2015-03-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for decreased induction of class IB PI3-kinases expression by MIF inhibitors.
J. Cell. Mol. Med., 21, 2017
|
|
4Z1U
 
 | | MIF in complex with 1-(4-methylphenyl)-3-phenylprop-2-yn-1-one | | Descriptor: | GLYCEROL, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Cho, T.Y. | | Deposit date: | 2015-03-27 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for decreased induction of class IB PI3-kinases expression by MIF inhibitors.
J. Cell. Mol. Med., 21, 2017
|
|
4UE0
 
 | |
3UIL
 
 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3UMQ
 
 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3USX
 
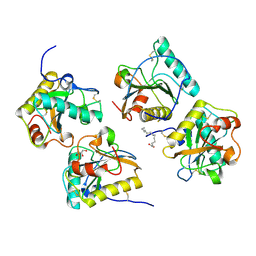 | | Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution | | Descriptor: | GLYCEROL, MYRISTIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Yamini, S, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3FAQ
 
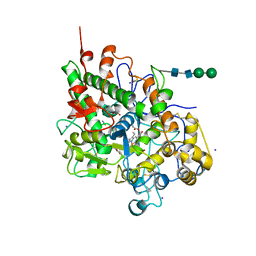 | | Crystal structure of lactoperoxidase complex with cyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
8D1B
 
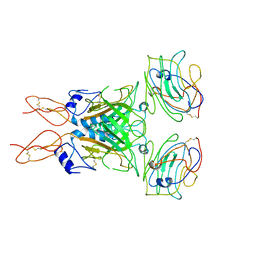 | |
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
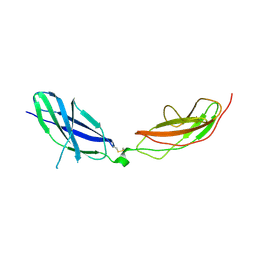 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
