1SS1
 
 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3AJI
 
 | | Structure of Gankyrin-S6ATPase photo-cross-linked site-specifically, and incoporated by genetic code expansion | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, Proteasome (Prosome, macropain) 26S subunit, ... | | Authors: | Sato, S, Mimasu, S, Sato, A, Hino, N, Sakamoto, K, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-07 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid
Biochemistry, 50, 2011
|
|
7D69
 
 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
5ZDN
 
 | | The complex structure of FomD with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, FomD, GLYCEROL, ... | | Authors: | Sato, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-02-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and Structural Analysis of FomD That Catalyzes the Hydrolysis of Cytidylyl ( S)-2-Hydroxypropylphosphonate in Fosfomycin Biosynthesis.
Biochemistry, 57, 2018
|
|
5ZDM
 
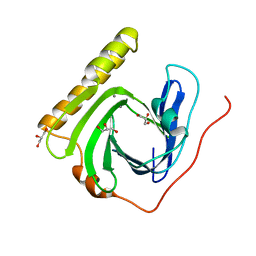 | | The ligand-free structure of FomD | | Descriptor: | CALCIUM ION, FomD, GLYCEROL | | Authors: | Sato, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-02-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biochemical and Structural Analysis of FomD That Catalyzes the Hydrolysis of Cytidylyl ( S)-2-Hydroxypropylphosphonate in Fosfomycin Biosynthesis.
Biochemistry, 57, 2018
|
|
3AWH
 
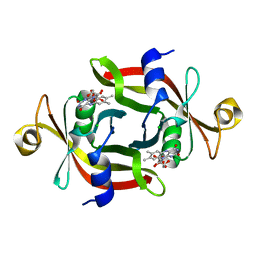 | |
3AMF
 
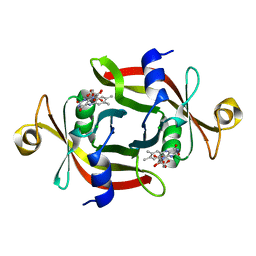 | |
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
1X3K
 
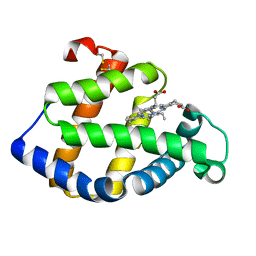 | | Crystal structure of a hemoglobin component (TA-V) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component V | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
1X46
 
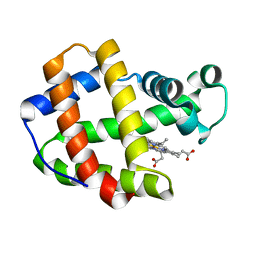 | | Crystal structure of a hemoglobin component (TA-VII) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component VII | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
8INL
 
 | | LSD1 in complex with S2172 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | LSD1 in complex with S2172
To Be Published
|
|
1W4K
 
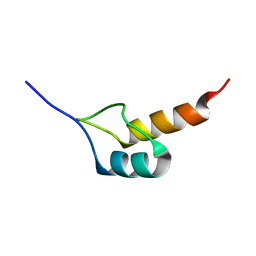 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4E
 
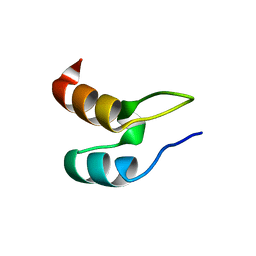 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4I
 
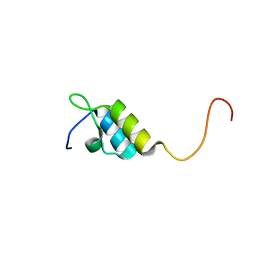 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
6KVD
 
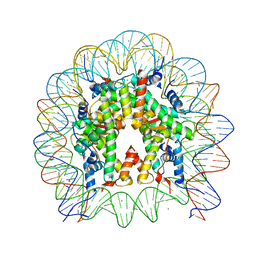 | | Crystal structure of human nucleosome containing H2A.J | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.J, ... | | Authors: | Tanaka, H, Koyama, M, Sato, S, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and structural analyses of the nucleosome containing human histone H2A.J.
J.Biochem., 167, 2020
|
|
5CPG
 
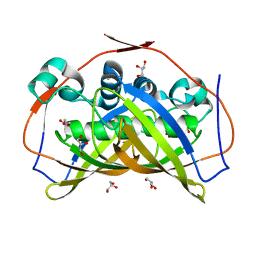 | | R-Hydratase PhaJ1 from Pseudomonas aeruginosa in the unliganded form | | Descriptor: | (R)-specific enoyl-CoA hydratase, GLYCEROL | | Authors: | Tsuge, T, Sato, S, Hiroe, A, Ishizuka, K, Kanazawa, H, Kanagarajan, S, Shiro, Y, Hisano, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Contribution of the Distal Pocket Residue to the Acyl-Chain-Length Specificity of (R)-Specific Enoyl-Coenzyme A Hydratases from Pseudomonas spp.
Appl.Environ.Microbiol., 81, 2015
|
|
5H6Q
 
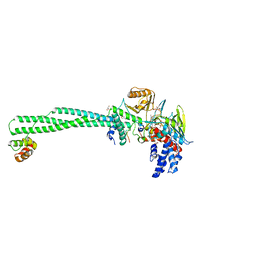 | | Crystal structure of LSD1-CoREST in complex with peptide 11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5H6R
 
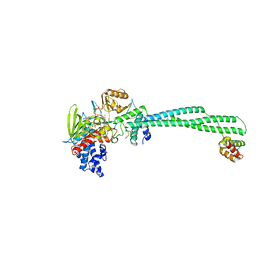 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
7XUA
 
 | | Structure of G9a in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUB
 
 | | Structure of G9a in complex with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUD
 
 | | Structure of G9a in complex with compound 26a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
8JW3
 
 | | The crystal structure of the viral terpene synthase from Orpheovirus IHUMI-LCC2 | | Descriptor: | SULFATE ION, Terpenoid synthase | | Authors: | Jung, Y, Mitsuhashi, T, Senda, M, Sato, S, Senda, T, Fujita, M. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Function and Structure of a Terpene Synthase Encoded in a Giant Virus Genome.
J.Am.Chem.Soc., 145, 2023
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
