6RKO
 
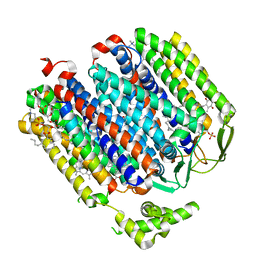 | | Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Safarian, S, Hahn, A, Kuehlbrandt, W, Michel, H. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Active site rearrangement and structural divergence in prokaryotic respiratory oxidases.
Science, 366, 2019
|
|
7NKZ
 
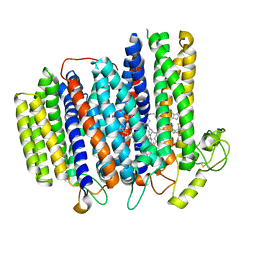 | | Cryo-EM structure of the cytochrome bd oxidase from M. tuberculosis at 2.5 A resolution | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, HEME B/C, MENAQUINONE-9, ... | | Authors: | Safarian, S, Wu, D, Krause, K.L, Michel, H. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The cryo-EM structure of the bd oxidase from M. tuberculosis reveals a unique structural framework and enables rational drug design to combat TB.
Nat Commun, 12, 2021
|
|
5IR6
 
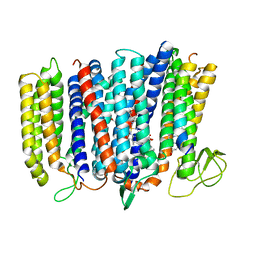 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2016-03-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
5DOQ
 
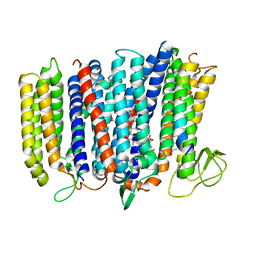 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2015-09-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
7OY2
 
 | | High resolution structure of cytochrome bd-II oxidase from E. coli | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(2~{E},6~{E},10~{Z},14~{E},18~{E},22~{E},26~{E})-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaenyl]naphthalene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Grund, T.N, Wu, D, Bald, D, Michel, H, Safarian, S. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Mechanistic and structural diversity between cytochrome bd isoforms of Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6HFB
 
 | | Outward-facing conformation of a multidrug resistance MATE family transporter of the MOP superfamily. | | Descriptor: | CESIUM ION, Uncharacterized protein | | Authors: | Nonaka, T, Zakrzewska, S, Safarian, S, Michel, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7ZDF
 
 | |
7ZDU
 
 | |
6YUZ
 
 | | Homodimeric structure of the rBAT complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YUP
 
 | | Heterotetrameric structure of the rBAT-b(0,+)AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT, ... | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YV1
 
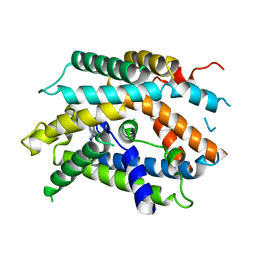 | | Structure of human b(0,+)AT1 | | Descriptor: | b(0,+)-type amino acid transporter 1 | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZDC
 
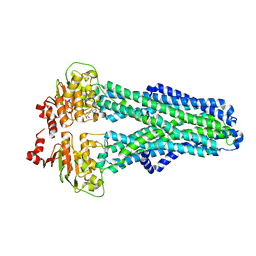 | |
7ZDA
 
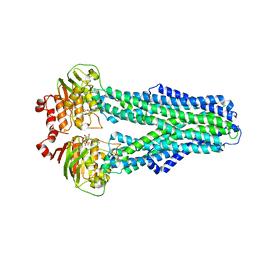 | |
7ZDK
 
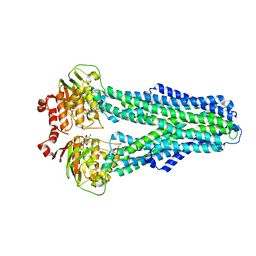 | |
7ZDV
 
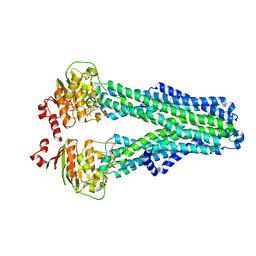 | |
7ZDB
 
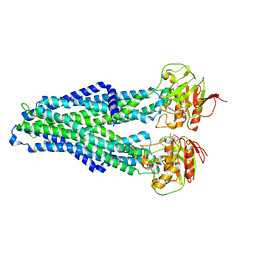 | |
7ZDE
 
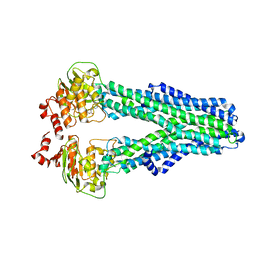 | |
7ZDG
 
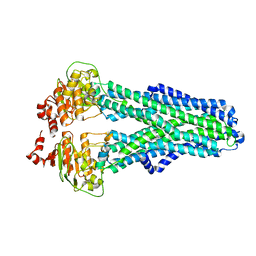 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDS
 
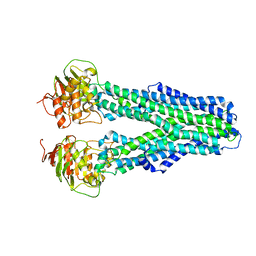 | |
7ZDL
 
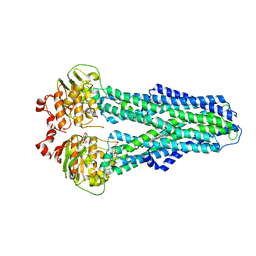 | |
7ZDR
 
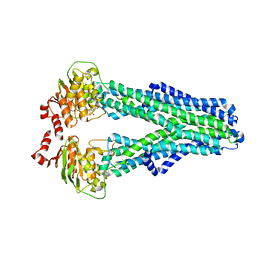 | |
7ZDW
 
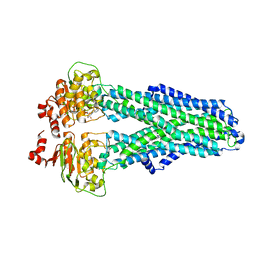 | |
7ZEC
 
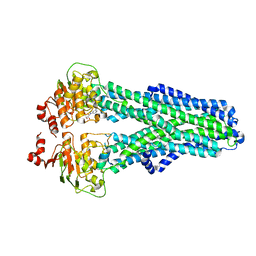 | |
7ZD5
 
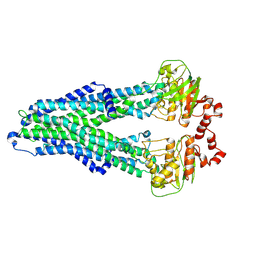 | | IF(apo/as isolated) conformation of CydDC (Dataset-1) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDT
 
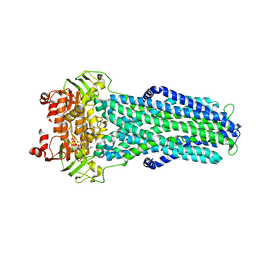 | |
