3TPK
 
 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NOM
 
 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3SI0
 
 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3SI2
 
 | | Structure of glycosylated murine glutaminyl cyclase in presence of the inhibitor PQ50 (PDBD150) | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3UJ1
 
 | |
7AQ1
 
 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
6GV1
 
 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
6SHT
 
 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
1YWJ
 
 | | Structure of the FBP11WW1 domain | | Descriptor: | Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1YWI
 
 | | Structure of the FBP11WW1 domain complexed to the peptide APPTPPPLPP | | Descriptor: | Formin, Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
6QRO
 
 | | Crystal structure of Tannerella forsythia glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
6QQL
 
 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
4NIW
 
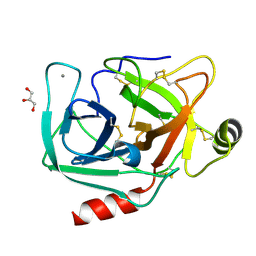 | |
4NIV
 
 | |
4NIY
 
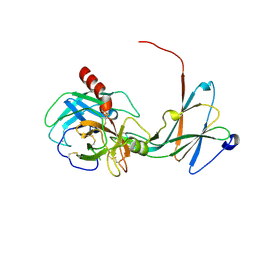 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) complexed to YRH-ecotin (M84Y/M85R/A86H ecotin) | | Descriptor: | CALCIUM ION, Cationic trypsin, Ecotin, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIX
 
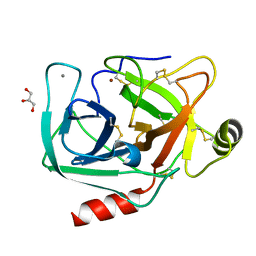 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3OE1
 
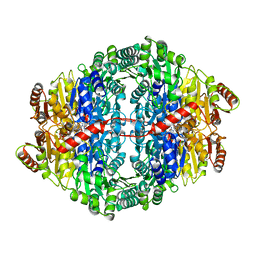 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
3S1X
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-sedoheptulose 7-phosphate Schiff-base intermediate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1U
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-erythrose 4-phosphate | | Descriptor: | CHLORIDE ION, ERYTHOSE-4-PHOSPHATE, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3SI1
 
 | | Structure of glycosylated murine glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Dambe, T, Carrillo, D, Parthier, C, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3S0C
 
 | | Transaldolase wt of Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1W
 
 | | Transaldolase variant Lys86Ala from Thermoplasma acidophilum in complex with glycerol and citrate | | Descriptor: | CITRATE ANION, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1V
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-fructose 6-phosphate Schiff-base intermediate | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
5C05
 
 | | Crystal Structure of Gamma-terpinene Synthase from Thymus vulgaris | | Descriptor: | 1,2-ETHANEDIOL, Putative gamma-terpinene synthase | | Authors: | Parthier, C.P, Rudolph, K, Muller, Y.A, Mueller-Uri, F. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Expression, crystallization and structure elucidation of gamma-terpinene synthase from Thymus vulgaris.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2R8P
 
 | | Transketolase from E. coli in complex with substrate D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium-2-yl}-6-O-phosphono-D-glucitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
