6EEY
 
 | |
6E58
 
 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6ELI
 
 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
7SZR
 
 | | NIK bound to inhibitor G02792917 | | Descriptor: | 1-(3-{[(1R,4R,5S)-4-hydroxy-2-methyl-3-oxo-2-azabicyclo[3.1.0]hexan-4-yl]ethynyl}phenyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Liau, N.P.D, Hymowitz, S.G. | | Deposit date: | 2021-11-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a nick in NIK: Extending the half-life of a NIK inhibitor through structure-based drug design.
Bioorg.Med.Chem.Lett., 89, 2023
|
|
7TUM
 
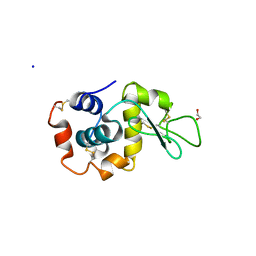 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7U6M
 
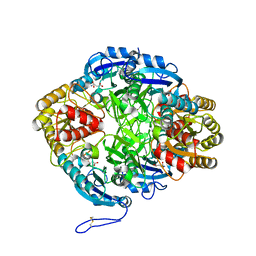 | |
7UEA
 
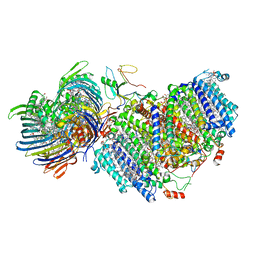 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
7UEB
 
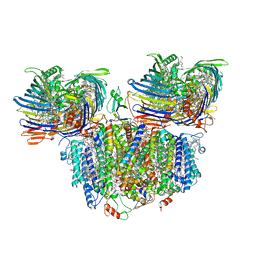 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
6FIB
 
 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7O5R
 
 | | Crystal structure of holo-SwHPA-Mn (hydroxyketoacid aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O9R
 
 | | Crystal structure of holo-H44A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-16 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5I
 
 | | Crystal structure of apo-SwHKA (Hydroxy ketone aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-08 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OBU
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1, with the active site in the resting and the active state | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5V
 
 | | Crystal structure of holo-H44A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1, in complex with Hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O87
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4WNI
 
 | |
4WNC
 
 | | Crystal structure of human wild-type GAPDH at 1.99 angstroms resolution | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Garcin, E.D. | | Deposit date: | 2014-10-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Dimer Interface Mutation in Glyceraldehyde-3-Phosphate Dehydrogenase Regulates Its Binding to AU-rich RNA.
J.Biol.Chem., 290, 2015
|
|
7OXL
 
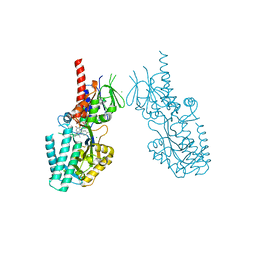 | | Crystal structure of human Spermine Oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Johannsson, S, Thomsen, M, Krapp, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
7OY0
 
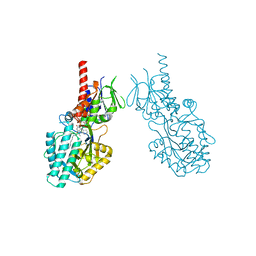 | | Structure of human Spermine Oxidase in complex with a highly selective allosteric inhibitor | | Descriptor: | 4-[(4-imidazo[1,2-a]pyridin-3-yl-1,3-thiazol-2-yl)amino]phenol, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Thomsen, M, Johannsson, S, Krapp, S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
4YIX
 
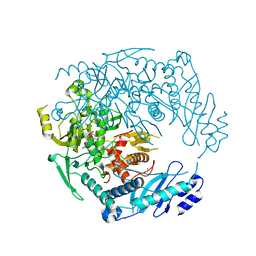 | | Structure of MRB1590 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
4YHJ
 
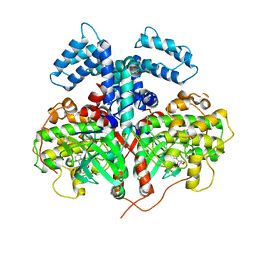 | | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4 (GRK4) | | Descriptor: | AMP PHOSPHORAMIDATE, G protein-coupled receptor kinase 4 | | Authors: | Allen, S.J, Parthasarathy, G, Soisson, S, Munshi, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4.
J.Biol.Chem., 290, 2015
|
|
4YIY
 
 | | Structure of MRB1590 bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, kRNA Editing A6 Specific Protein | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.016 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
7KF6
 
 | |
7KF7
 
 | |
