1LA2
 
 | | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase | | Descriptor: | Myo-inositol-1-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kniewel, R, Buglino, J.A, Shen, V, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-10 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1P1L
 
 | |
1P1M
 
 | | Structure of Thermotoga maritima amidohydrolase TM0936 bound to Ni and methionine | | Descriptor: | Hypothetical protein TM0936, METHIONINE, NICKEL (II) ION | | Authors: | Kniewel, R, Buglino, J.A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the hypothetical protein TM0936 from Thermotoga maritima at
1.5A bound to Ni and methionine
To be Published, 2003
|
|
1PUJ
 
 | | Structure of B. subtilis YlqF GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, conserved hypothetical protein ylqF | | Authors: | Kniewel, R, Buglino, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YlqF GTPase from B. subtilis
To be Published
|
|
1PUG
 
 | | Structure of E. coli Ybab | | Descriptor: | Hypothetical UPF0133 protein ybaB | | Authors: | Kniewel, R, Buglino, J, Chadna, T, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of E. coli Ybab
To be Published
|
|
1PUI
 
 | |
1PSQ
 
 | | Structure of a probable thiol peroxidase from Streptococcus pneumoniae | | Descriptor: | probable thiol peroxidase | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a probable thiol peroxidase from Streptococcus pneumoniae
To be Published
|
|
1PSW
 
 | | Structure of E. coli ADP-heptose lps heptosyltransferase II | | Descriptor: | ADP-HEPTOSE LPS HEPTOSYLTRANSFERASE II | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of E. coli ADP-heptose lps heptosyltransferase II
To be Published
|
|
1Q98
 
 | | Structure of a Thiol Peroxidase from Haemophilus influenzae Rd | | Descriptor: | Thiol Peroxidase | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-09 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Thiol Peroxidase from Haemophilus influenzae Rd
To be Published
|
|
1TLQ
 
 | | Crystal structure of protein ypjQ from Bacillus subtilis, Pfam DUF64 | | Descriptor: | CALCIUM ION, Hypothetical protein ypjQ | | Authors: | Kniewel, R, Rajashankar, K.R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Hypothetical Protein from Bacillus subtilis
To be Published
|
|
2FS2
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
1PSU
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein PaaI | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
4LZ6
 
 | | Structure of MATE multidrug transporter DinF-BH | | Descriptor: | BH2163 protein | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LZ9
 
 | | Structure of MATE multidrug transporter DinF-BH in complex with R6G | | Descriptor: | BH2163 protein, RHODAMINE 6G | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
1LX7
 
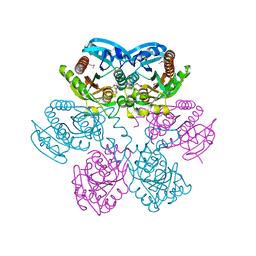 | | Structure of E. coli uridine phosphorylase at 2.0A | | Descriptor: | uridine phosphorylase | | Authors: | Burling, T, Buglino, J.A, Kniewel, R, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-04 | | Release date: | 2002-06-12 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli uridine phosphorylase at 2.0 A.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6YYI
 
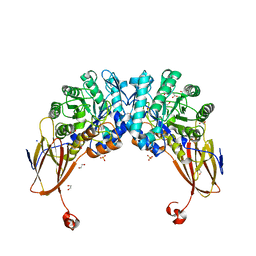 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
6YYH
 
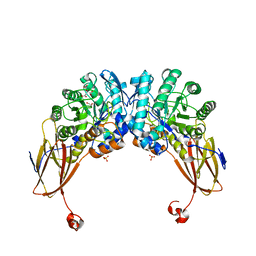 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
2A8X
 
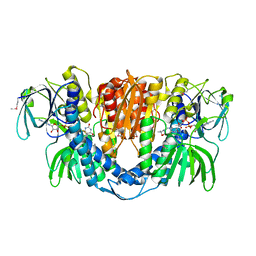 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
6I60
 
 | | Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Alpha-rhamnosidase, TRIETHYLENE GLYCOL | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Biochemical Characterization of the alpha-l-RhamnosidaseDtRha fromDictyoglomus thermophilum: Application to the Selective Derhamnosylation of Natural Flavonoids.
Acs Omega, 4, 2019
|
|
6F3K
 
 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6G7O
 
 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
8CLR
 
 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-09 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
6XXW
 
 | | Structure of beta-D-Glucuronidase for Dictyoglomus thermophilum. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucuronidase, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Thioglycoligation of aromatic thiols using a natural glucuronide donor.
Org.Biomol.Chem., 18, 2020
|
|
2CMU
 
 | |
1Z9D
 
 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | Descriptor: | SULFATE ION, uridylate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
