1SDE
 
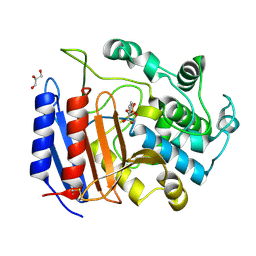 | | Toward Better Antibiotics: Crystal Structure Of D-Ala-D-Ala Peptidase inhibited by a novel bicyclic phosphate inhibitor | | Descriptor: | 2-[(DIOXIDOPHOSPHINO)OXY]BENZOATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-13 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward better antibiotics: crystallographic studies of a novel class of DD-peptidase/beta-lactamase inhibitors
Biochemistry, 43, 2004
|
|
2XDV
 
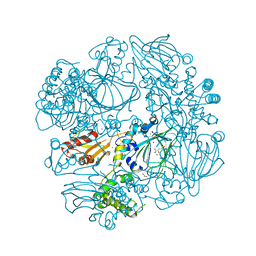 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | Authors: | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-07 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2XGN
 
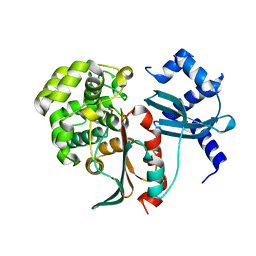 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-07 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFT
 
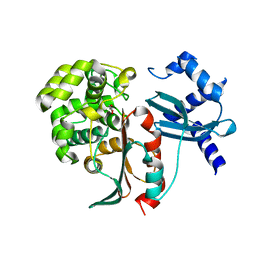 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XH9
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XF3
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFS
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
5LAS
 
 | |
5LB6
 
 | |
5LBE
 
 | |
5LBC
 
 | |
5LBB
 
 | |
5L9R
 
 | |
5L9B
 
 | |
5LBF
 
 | |
5LA9
 
 | |
5LAT
 
 | |
5MMY
 
 | | Crystal structure of OXA10 with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase OXA-10, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5MOX
 
 | | OXA-10 Avibactam complex with bound CO2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, CARBON DIOXIDE, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5MOZ
 
 | | OXA-10 Avibactam complex with bound Iodide | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, CHLORIDE ION, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5N5H
 
 | | Crystal structure of metallo-beta-lactamase VIM-1 in complex with ML302F inhibitor | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5N5I
 
 | | Crystal Structure of VIM-1 metallo-beta-lactamase in complex with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
4AI9
 
 | | JMJD2A Complexed with Daminozide | | Descriptor: | CHLORIDE ION, DAMINOZIDE, GLYCEROL, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases.
J. Med. Chem., 55, 2012
|
|
4B7K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4BIS
 
 | | JMJD2A COMPLEXED WITH 8-HYDROXYQUINOLINE-4-CARBOXYLIC ACID | | Descriptor: | 8-hydroxyquinoline-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Thinnes, C, Schofield, C.J. | | Deposit date: | 2013-04-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation.
Chem Sci, 4, 2013
|
|
