2J5P
 
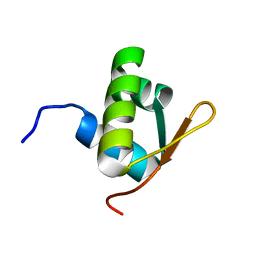 | | E. coli FtsK gamma domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Sivanathan, V, Allen, M.D, de Bekker, C, Baker, R, Arciszewska, L, Freund, S.M, Bycroft, M, Lowe, J, Sherratt, D.J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ftsk Gamma Domain Directs Oriented DNA Translocation by Interacting with Kops.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5MN4
 
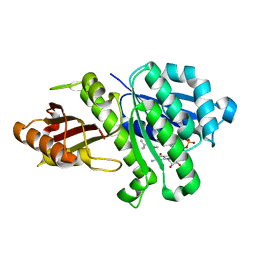 | | S. aureus FtsZ 12-316 F138A GDP Open form (1FOf) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN8
 
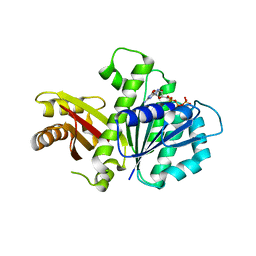 | | S. aureus FtsZ 12-316 F138A GTP Closed form (5FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MW1
 
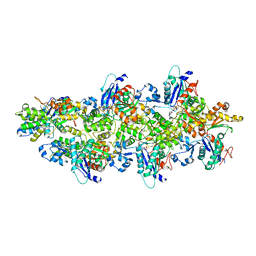 | |
5MN5
 
 | | S. aureus FtsZ 12-316 T66W GTP Closed form (2TCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN6
 
 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN7
 
 | | S. aureus FtsZ 12-316 F138A GTP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5N8P
 
 | |
5N97
 
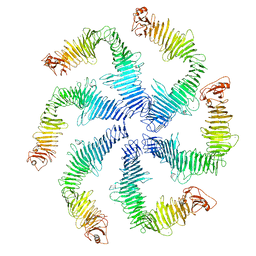 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
5NR1
 
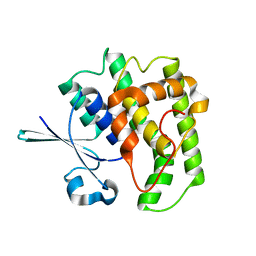 | | FzlA from C. crescentus | | Descriptor: | FtsZ-binding protein FzlA | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division.
Mol. Microbiol., 107, 2018
|
|
5O09
 
 | | BtubABC mini microtubule | | Descriptor: | Bacterial kinesin light chain, GUANOSINE-5'-DIPHOSPHATE, Tubulin, ... | | Authors: | Deng, X, Bharat, T.A.M, Lowe, J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1OI0
 
 | | CRYSTAL STRUCTURE OF AF2198, A JAB1/MPN DOMAIN PROTEIN FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | HYPOTHETICAL PROTEIN AF2198, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ZINC ION | | Authors: | Tran, H.J.T.T, Allen, M.D, Lowe, J, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-08-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Jab1/Mpn Domain and its Implications for Proteasome Function
Biochemistry, 42, 2003
|
|
1OFT
 
 | |
1OFU
 
 | |
1JCE
 
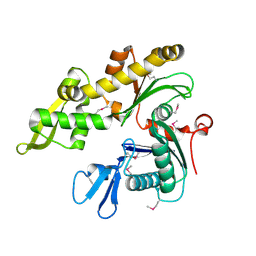 | |
1JCG
 
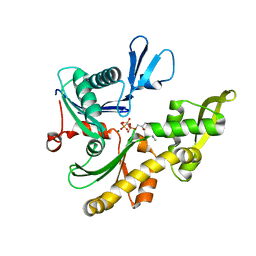 | | MREB FROM THERMOTOGA MARITIMA, AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | van den Ent, F, Amos, L.A, Lowe, J. | | Deposit date: | 2001-06-09 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prokaryotic origin of the actin cytoskeleton.
Nature, 413, 2001
|
|
1JCF
 
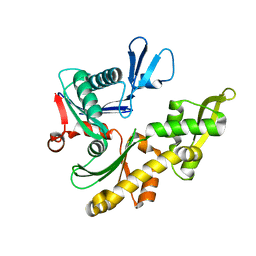 | |
1PMA
 
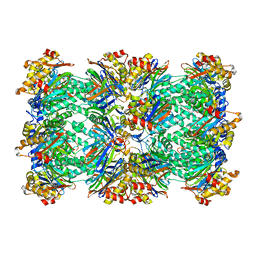 | | PROTEASOME FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PROTEASOME | | Authors: | Loewe, J, Stock, D, Jap, B, Zwickl, P, Baumeister, W, Huber, R. | | Deposit date: | 1994-12-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Science, 268, 1995
|
|
8DKN
 
 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DKV
 
 | | PPARg bound to JTP-426467 and Co-R peptide | | Descriptor: | 2-chloro-N-[4-(5-methyl-1,3-benzoxazol-2-yl)phenyl]-5-nitrobenzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSY
 
 | | PPARg bound to inverse agonist H3B-343 | | Descriptor: | Peroxisome proliferator-activated receptor gamma, {5-[(5-{[(4-tert-butylphenyl)methyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}acetic acid | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
1A6E
 
 | | THERMOSOME-MG-ADP-ALF3 COMPLEX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
1A6D
 
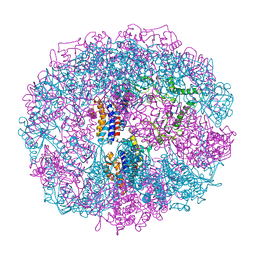 | | THERMOSOME FROM T. ACIDOPHILUM | | Descriptor: | THERMOSOME (ALPHA SUBUNIT), THERMOSOME (BETA SUBUNIT) | | Authors: | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
4K7P
 
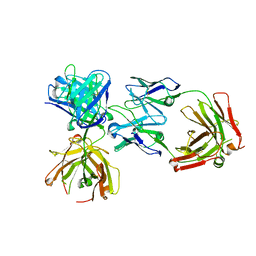 | |
