3WL8
 
 | | Crystal Structure of pOPH S172A with octanoic acid | | Descriptor: | CITRIC ACID, OCTANOIC ACID (CAPRYLIC ACID), Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WWE
 
 | | The complex of pOPH with PEG | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3WLA
 
 | | Crystal Structure of sOPH Native | | Descriptor: | Oxidized polyvinyl alcohol hydrolase, SULFATE ION | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WL7
 
 | | The complex structure of pOPH S172C with ligand, ACA | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase, pentane-2,4-dione | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WWC
 
 | | The complex of pOPH_S172A of pNPB | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase, butanoic acid | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3WWD
 
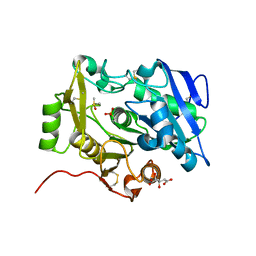 | | The complex of pOPH_S172C with DMSO | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
5HGZ
 
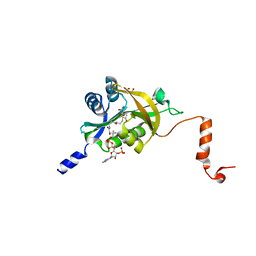 | | Crystal structure of human Naa60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, MALONIC ACID, N-alpha-acetyltransferase 60 | | Authors: | Chen, J.Y, Liu, L, Yun, C.H. | | Deposit date: | 2016-01-09 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase
Sci Rep, 6, 2016
|
|
5HH0
 
 | |
5IWZ
 
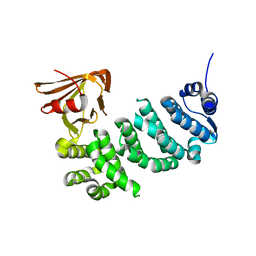 | | Synaptonemal complex protein | | Descriptor: | Synaptonemal complex protein 2 | | Authors: | Feng, J, Fu, S, Cao, X, Wu, H, Lu, J, Zeng, M, Liu, L, Yang, X, Shen, Y. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structure of synaptonemal complexes protein at 2.6 angstroms resolution
To Be Published
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
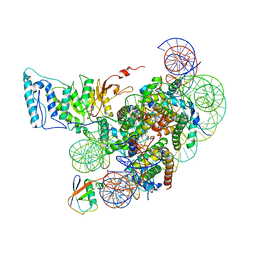 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
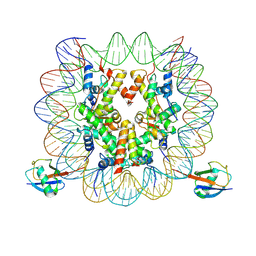 | | cryo-EM structure of H2BK34ub nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
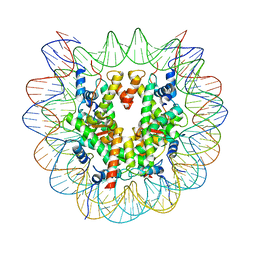 | | cryo-EM structure of unmodified nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
3WL5
 
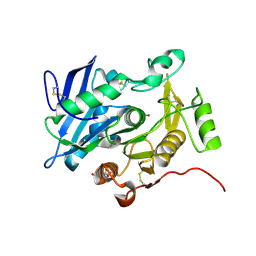 | | Crystal structure of pOPH S172C | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-07 | | Release date: | 2014-09-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
6JKV
 
 | | PppA, a key regulatory component of T6SS in Pseudomonas aeruginosa | | Descriptor: | MANGANESE (II) ION, PppA | | Authors: | Wang, T, Liu, L, Wu, Y, Li, D. | | Deposit date: | 2019-03-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
3WL6
 
 | | Crystal Structure of pOPH Native | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
5VBT
 
 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
8GCM
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCP
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4ZDS
 
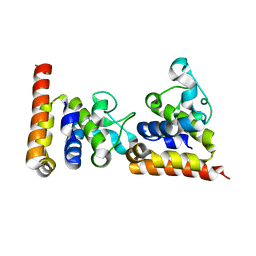 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
4ZHJ
 
 | |
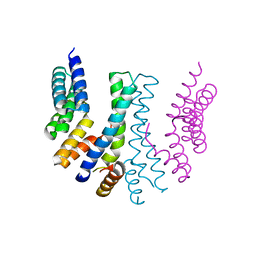
4YVQ
 
 | |
5B6L
 
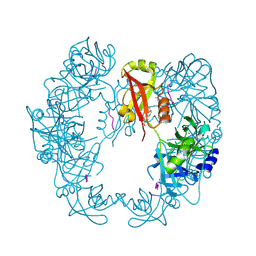 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
5CHE
 
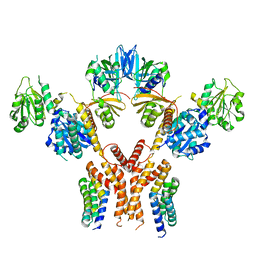 | |
5FEJ
 
 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
