8ILQ
 
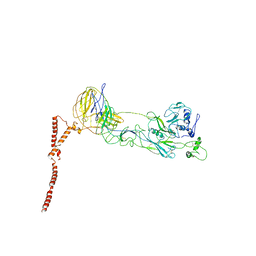 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
4RVP
 
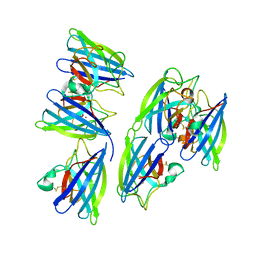 | | Crystal structure of superoxide dismutase from sedum alfredii | | Descriptor: | ZINC ION, superoxide dismutase | | Authors: | Qiu, R, Li, C, Zhai, J, Tang, L, Zhang, H, Yuan, M, Hu, X. | | Deposit date: | 2014-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The positive effects of Cd and Cd-Zn relationship in the
Zn-related physiological processes involved in growth in the Zn/Cd hyperaccumulator Sedum alfredii
To be Published
|
|
1JXS
 
 | | Solution Structure of the DNA-Binding Domain of Interleukin Enhancer Binding Factor | | Descriptor: | interleukin enhancer binding factor | | Authors: | Chuang, W.J, Liu, P.P, Li, C, Hsieh, Y.H, Chen, S.W, Chen, S.H, Jeng, W.Y. | | Deposit date: | 2001-09-08 | | Release date: | 2003-03-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
1KBK
 
 | | Mechanistic Analyses of Catalysis in Human Pancreatic Alpha-Amylase: Detailed Kinetic and Structural Studies of Mutants of Three Conserved Carboxylic Acids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Rydberg, E.H, Li, C, Maurus, R, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids.
Biochemistry, 41, 2002
|
|
1KBB
 
 | | Mechanistic Analyses of Catalysis in Human Pancreatic alpha-Amylase: Detailed Kinetic and Structural Studies of Mutants of Three Conserved Carboxylic Acids | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Li, C, Maurus, R, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-05 | | Release date: | 2002-04-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids.
Biochemistry, 41, 2002
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2IU3
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
6UGH
 
 | |
8D1T
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
1THZ
 
 | | Crystal Structure of Avian AICAR Transformylase in Complex with a Novel Inhibitor Identified by Virtual Ligand Screening | | Descriptor: | 2-{(E)-[5-HYDROXY-3-METHYL-1-(2-METHYL-4-SULFOPHENYL)-1H-PYRAZOL-4-YL]DIAZENYL}-4-SULFOBENZOIC ACID, Bifunctional purine biosynthesis protein PURH, POTASSIUM ION | | Authors: | Xu, L, Li, C, Olson, A.J, Wilson, I.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
J.Biol.Chem., 279, 2004
|
|
1U2Y
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U33
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-O-METHYL-MALTOSYL-ALPHA (1,4)-(Z, 3S,4S,5R,6R)-3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-PIPERIDIN-2-ONE, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U30
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1YSW
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1NJS
 
 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1PW7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-30 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PK7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with adenosine and sulfate/phosphate | | Descriptor: | ADENOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PK9
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR4
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
