1FC3
 
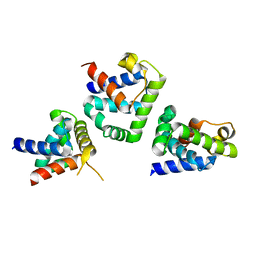 | |
1QMP
 
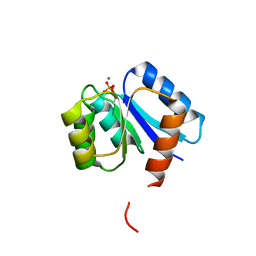 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
1B0N
 
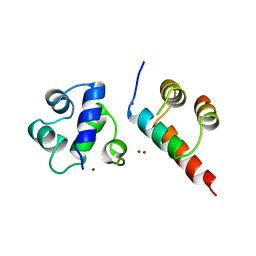 | | SINR PROTEIN/SINI PROTEIN COMPLEX | | Descriptor: | PROTEIN (SINI PROTEIN), PROTEIN (SINR PROTEIN), ZINC ION | | Authors: | Lewis, R.J, Brannigan, J.A, Offen, W.A, Smith, I, Wilkinson, A.J. | | Deposit date: | 1998-11-11 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J.Mol.Biol., 283, 1998
|
|
1DZ3
 
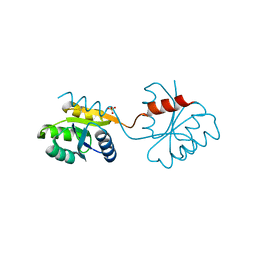 | | DOMAIN-SWAPPING IN THE SPORULATION RESPONSE REGULATOR SPO0A | | Descriptor: | SULFATE ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Leonard, G, Barak, I, Wilkinson, A.J. | | Deposit date: | 2000-02-15 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Domain swapping in the sporulation response regulator Spo0A.
J. Mol. Biol., 297, 2000
|
|
6GQA
 
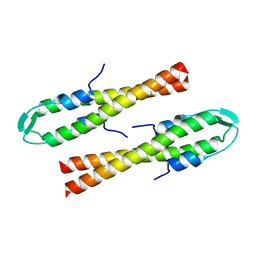 | | Cell division regulator S. pneumoniae GpsB | | Descriptor: | Cell cycle protein GpsB | | Authors: | Lewis, R.J, Rutter, Z.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes.
Nat Commun, 10, 2019
|
|
6GQN
 
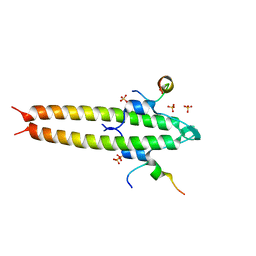 | | Cell division regulator, S. pneumoniae GpsB, in complex with peptide fragment of Penicillin Binding Protein PBP2a | | Descriptor: | Cell cycle protein GpsB, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Lewis, R.J, Rutter, Z.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes.
Nat Commun, 10, 2019
|
|
4UXV
 
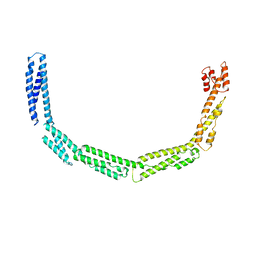 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4UY3
 
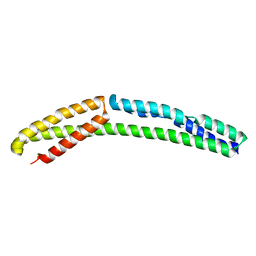 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
1W53
 
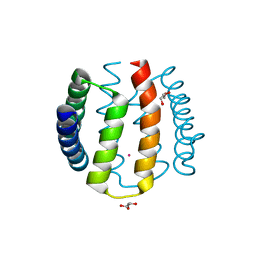 | | Kinase recruitment domain of the stress phosphatase RsbU | | Descriptor: | GLYCEROL, PHOSPHOSERINE PHOSPHATASE RSBU, XENON | | Authors: | Delumeau, O, Dutta, S, Brigulla, M, Kuhnke, G, Hardwick, S.W, Voelker, U, Yudkin, M.D, Lewis, R.J. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Characterization of Rsbu, a Stress Signaling Protein Phosphatase 2C
J.Biol.Chem., 279, 2004
|
|
1MVI
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA, NMR, 15 STRUCTURES | | Descriptor: | MVIIA | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1MVJ
 
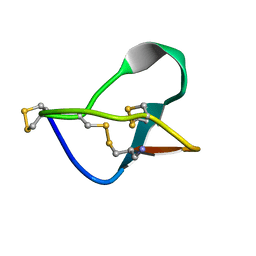 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1MTQ
 
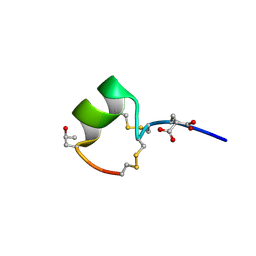 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID BY NMR SPECTROSCOPY | | Descriptor: | alpha-conotoxin GID | | Authors: | Nicke, A, Loughnan, M.L, Millard, E.L, Alewood, P.F, Adams, D.J, Daly, N.L, Craik, D.J, Lewis, R.J. | | Deposit date: | 2002-09-22 | | Release date: | 2003-02-11 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Isolation, Structure, and Activity of GID, a Novel alpha 4/7-Conotoxin with an Extended N-terminal Sequence
J.BIOL.CHEM., 278, 2003
|
|
7BN9
 
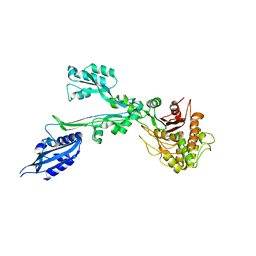 | |
7SKC
 
 | |
7O61
 
 | |
6GP7
 
 | |
6GPZ
 
 | | Cell division regulator GpsB in complex with peptide fragment of L. monocytogenes Penicillin Binding Protein PBPA1 | | Descriptor: | Cell cycle protein GpsB, IMIDAZOLE, LmPBPA1, ... | | Authors: | Cleverley, R.M, Lewis, R.J, Rutter, Z.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes.
Nat Commun, 10, 2019
|
|
5OMT
 
 | | Endonuclease NucB | | Descriptor: | NucB | | Authors: | Basle, A, Lewis, R.J. | | Deposit date: | 2017-08-01 | | Release date: | 2017-11-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of NucB, a biofilm-degrading endonuclease.
Nucleic Acids Res., 46, 2018
|
|
7TXF
 
 | |
3K92
 
 | | Crystal structure of a E93K mutant of the majour Bacillus subtilis glutamate dehydrogenase RocG | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
3K8Z
 
 | | Crystal Structure of Gudb1 a decryptified secondary glutamate dehydrogenase from B. subtilis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
1FSE
 
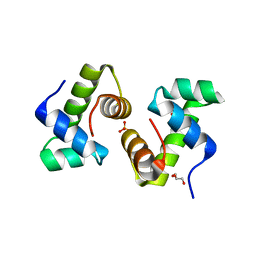 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
4HHF
 
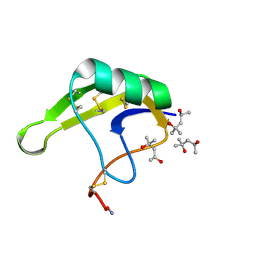 | | Crystal Structure of chemically synthesized scorpion alpha-toxin OD1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-toxin OD1 | | Authors: | Wang, C.I.A, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-10-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical engineering and structural and pharmacological characterization of the alpha-scorpion toxin OD1.
Acs Chem.Biol., 8, 2013
|
|
6TIF
 
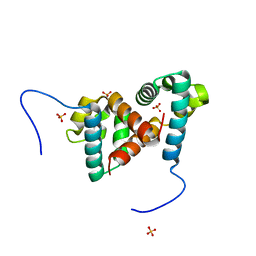 | | ReoM- Listeria monocytogenes | | Descriptor: | SULFATE ION, UPF0297 protein lmo1503 | | Authors: | Rutter, Z.J, Lewis, R.J. | | Deposit date: | 2019-11-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM.
Elife, 9, 2020
|
|
4I6O
 
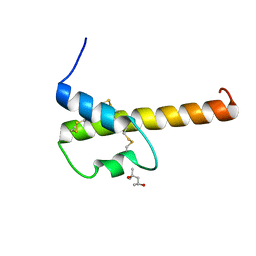 | | Crystal structure of chemically synthesized human anaphylatoxin C3a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Complement C3 | | Authors: | Wang, C.I.A, Ghassemian, A, Collins, B, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-27 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Efficient chemical synthesis of human complement protein C3a.
Chem.Commun.(Camb.), 49, 2013
|
|
