3AFA
 
 | | The human nucleosome structure | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AN2
 
 | | The structure of the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Kagawa, W, Shiga, T, Saito, K, Osakabe, A, Hayashi-Takanaka, Y, Park, S.-Y, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-08-27 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the human centromeric nucleosome containing CENP-A
Nature, 476, 2011
|
|
3AV1
 
 | | The human nucleosome structure containing the histone variant H3.2 | | Descriptor: | 146-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Osakabe, A, Shiga, T, Miya, Y, Kimura, H, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human nucleosomes containing major histone H3 variants
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AV2
 
 | | The human nucleosome structure containing the histone variant H3.3 | | Descriptor: | 146-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Osakabe, A, Shiga, T, Miya, M, Kimura, H, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human nucleosomes containing major histone H3 variants
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7VBM
 
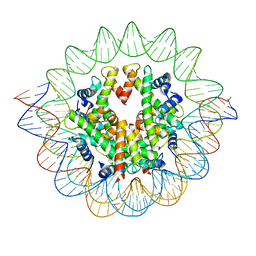 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
7VZ4
 
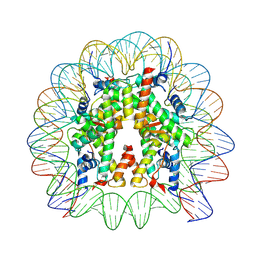 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601L DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Sato, S, Danev, R, Kurumizaka, H. | | Deposit date: | 2021-11-15 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Methods for High Resolution Cryo-EM Analyses of Nucleosomes
To Be Published
|
|
7W9V
 
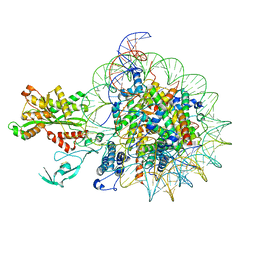 | | Cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex I) | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Hatazawa, S, Liu, J, Takizawa, Y, Zandian, M, Negishi, L, Kutateladze, T.G, Kurumizaka, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for binding diversity of acetyltransferase p300 to the nucleosome.
Iscience, 25, 2022
|
|
7X58
 
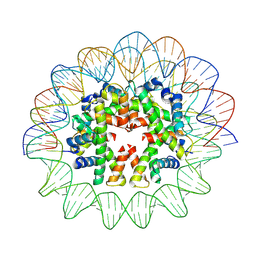 | | Cryo-EM structure of human subnucleosome (open form) | | Descriptor: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X57
 
 | | Cryo-EM structure of human subnucleosome (closed form) | | Descriptor: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A5L
 
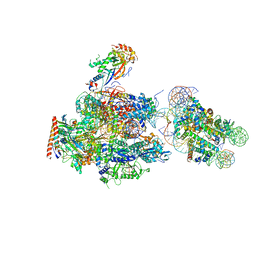 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5R
 
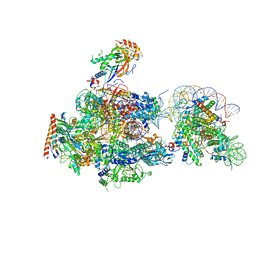 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5T
 
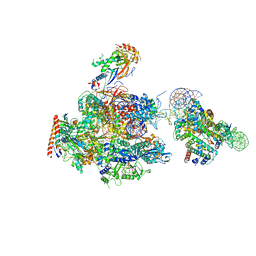 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5P
 
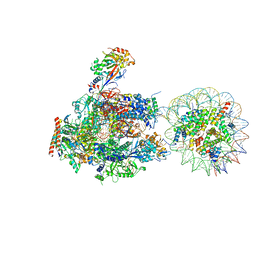 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5U
 
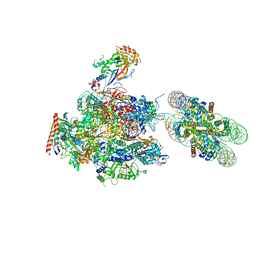 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
7XZX
 
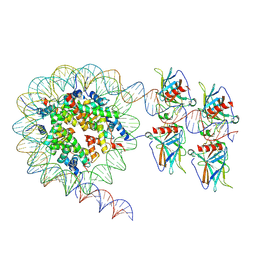 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZZ
 
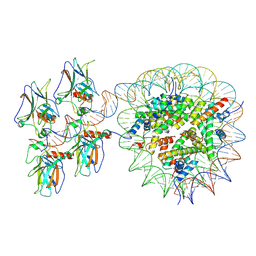 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZY
 
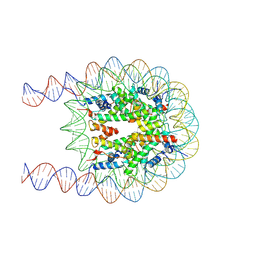 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7Y00
 
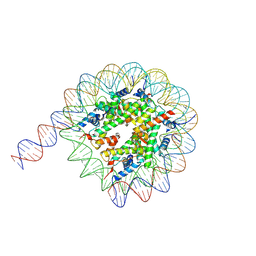 | | Cryo-EM structure of the nucleosome containing 169 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (169-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7YOZ
 
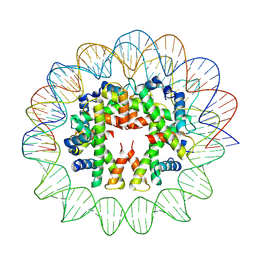 | | Cryo-EM structure of human subnucleosome (intermediate form) | | Descriptor: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A5O
 
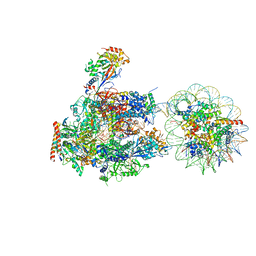 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
5XRZ
 
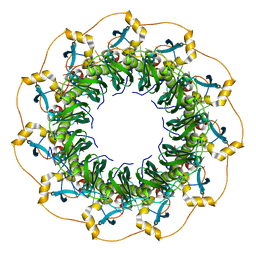 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
5XS0
 
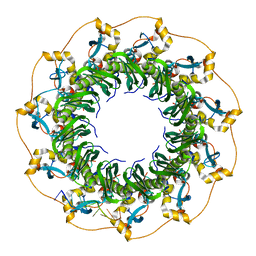 | | Structure of a ssDNA bound to the outer DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, ssDNA (5'-D(*CP*CP*CP*CP*CP*C)-3'), ssDNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
5B40
 
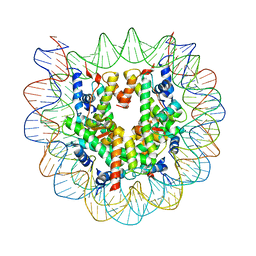 | | The nucleosome structure containing H2B-K120 and H4-K31 monoubiquitinations | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Machida, S, Sekine, S, Nishiyama, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
To Be Published
|
|
5B0Y
 
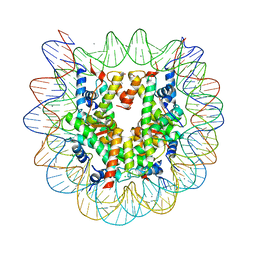 | | Crystal structure of the nucleosome containing histone H3 with the crotonylated lysine 122 | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Suzuki, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5B0Z
 
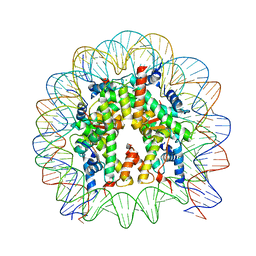 | | The crystal structure of the nucleosome containing H3.2, at 1.98 A resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Suzuki, Y, Horikoshi, N, Kato, D, Kurumizaka, H. | | Deposit date: | 2015-11-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
