2QHO
 
 | |
1L1P
 
 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
4EEW
 
 | |
5V8Z
 
 | |
5V90
 
 | |
3EC3
 
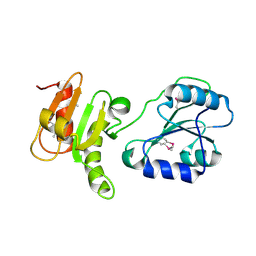 | | Crystal structure of the bb fragment of ERp72 | | Descriptor: | Protein disulfide-isomerase A4 | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2008-08-28 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Noncatalytic Domains and Global Fold of the Protein Disulfide Isomerase ERp72.
Structure, 17, 2009
|
|
3KTR
 
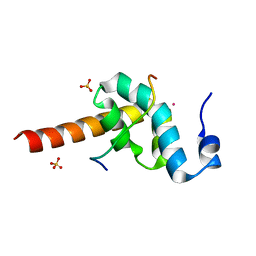 | |
3KTP
 
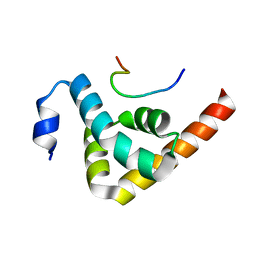 | | Structural basis of GW182 recognition by poly(A)-binding protein | | Descriptor: | Polyadenylate-binding protein 1, Trinucleotide repeat-containing gene 6C protein | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein.
J.Biol.Chem., 285, 2010
|
|
3KUT
 
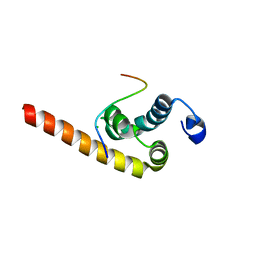 | |
3KUJ
 
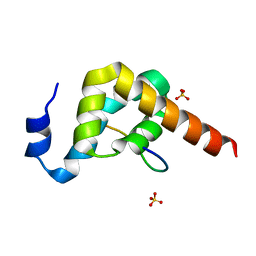 | |
3KUS
 
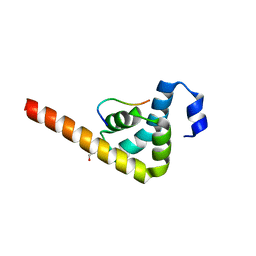 | |
3KUI
 
 | |
3KUR
 
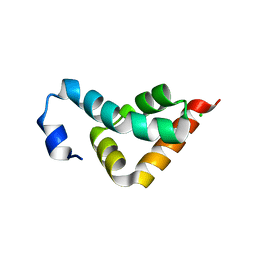 | |
6WUR
 
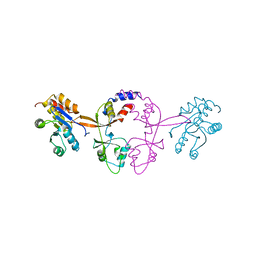 | |
6WUS
 
 | |
8G91
 
 | |
8G90
 
 | |
8EY8
 
 | |
8EY6
 
 | |
8EY7
 
 | |
1D5G
 
 | |
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1JH4
 
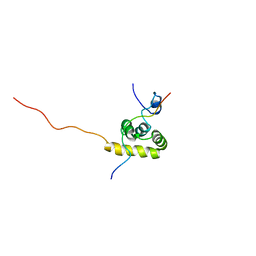 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
3PDZ
 
 | |
3GZH
 
 | |
