6A6E
 
 | | Crystal structure of thermostable Cysteine desulfurase (FiSufS) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | CITRIC ACID, Cysteine desulfurase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6A6F
 
 | | Crystal structure of Putative iron-sulfur cluster assembly scaffold protein for SUF system (FiSufU) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Iron-sulfur cluster assembly scaffold protein NifU, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6USF
 
 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6UR8
 
 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylcholine receptor subunit alpha-4, Neuronal acetylcholine receptor subunit beta-2, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
4ME2
 
 | | Crystal Structure of THA8 protein from Brachypodium distachyon | | Descriptor: | Uncharacterized protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Melcher, K, Xu, H.E. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2FWZ
 
 | |
6L48
 
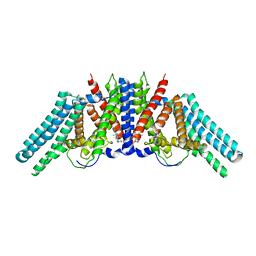 | |
6L47
 
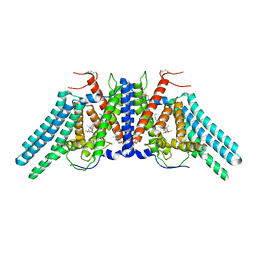 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
2FWY
 
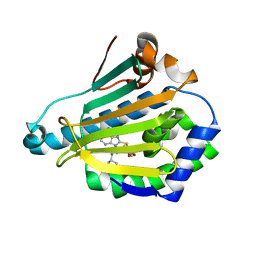 | |
2H55
 
 | |
5YKF
 
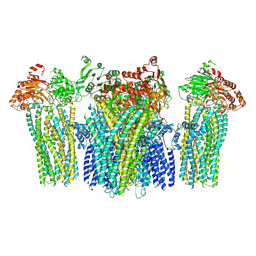 | |
5YKG
 
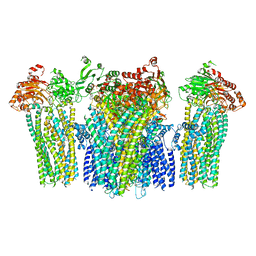 | |
5YKE
 
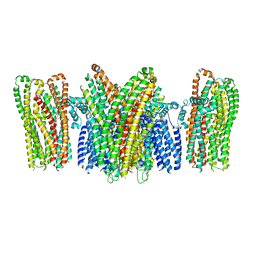 | |
6CBV
 
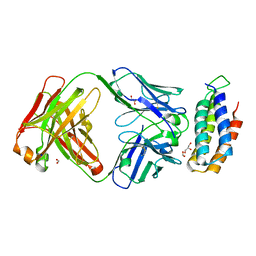 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
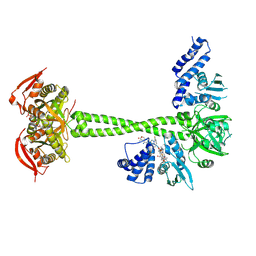
8HBE
 
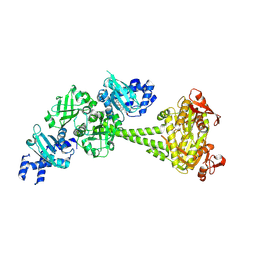 | |
8HBF
 
 | |
8HBH
 
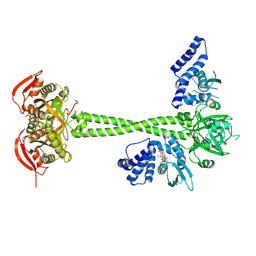 | |
7E4T
 
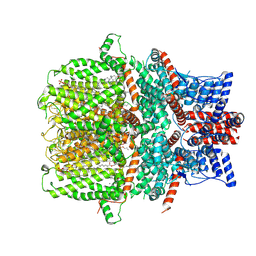 | | Human TRPC5 apo state structure at 3 angstrom | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7DXD
 
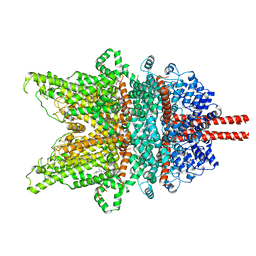 | | Structure of TRPC3 at 3.9 angstrom in 1340 nM free calcium state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Short transient receptor potential channel 3, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXC
 
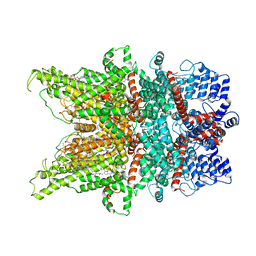 | | Structure of TRPC3 at 3.06 angstrom in low calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, Short transient receptor potential channel 3, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXF
 
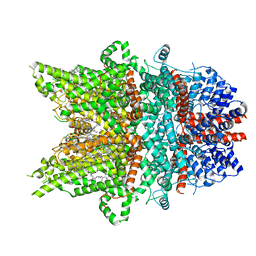 | | Structure of BTDM-bound human TRPC6 nanodisc at 2.9 angstrom in high calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXG
 
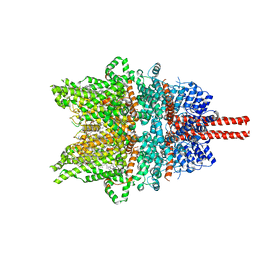 | | Structure of SAR7334-bound TRPC6 at 2.9 angstrom | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[[(1R,2R)-2-[(3R)-3-azanylpiperidin-1-yl]-2,3-dihydro-1H-inden-1-yl]oxy]-3-chloranyl-benzenecarbonitrile, CALCIUM ION, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7DXE
 
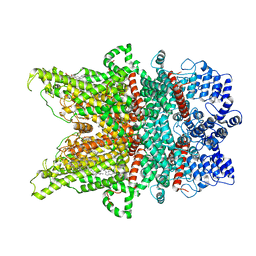 | |
7DXB
 
 | | Structure of TRPC3 at 2.7 angstrom in high calcium state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Guo, W. | | Deposit date: | 2021-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites.
Neuron, 110, 2022
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
