1BTK
 
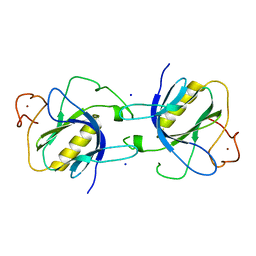 | |
6YJF
 
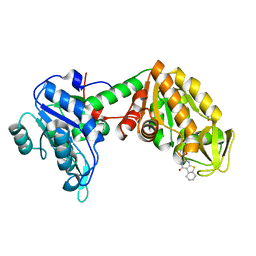 | | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEGSmear at pH 6.5 | | Descriptor: | (2~{S})-2-(5-nitrofuran-2-yl)-2,3,5,6,7,8-hexahydro-1~{H}-[1]benzothiolo[2,3-d]pyrimidin-4-one, GLYCEROL, Phosphoglycerate kinase | | Authors: | Hyvonen, M, Brear, P, Blaszczyk, B.K. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phosphoglycerate Kinase as a potential target for antimalarial therapy
to be published
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
8B9P
 
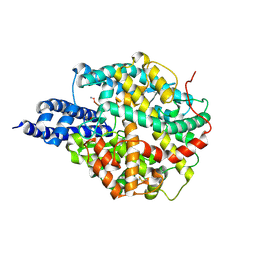 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
4UQO
 
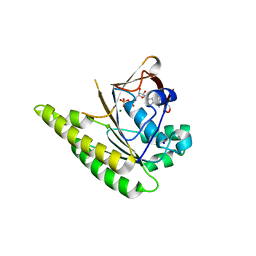 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-06-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
6Q62
 
 | |
7QUX
 
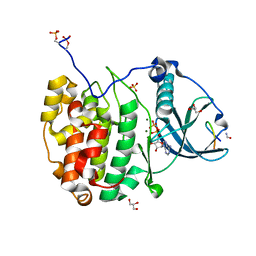 | | Crystal structure of P7C8 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2022-01-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
7NUF
 
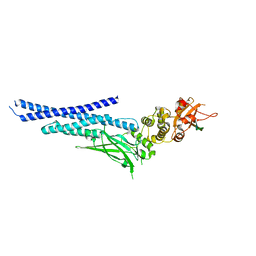 | | Vaccinia virus protein 018 in complex with STAT1 | | Descriptor: | ACETYL GROUP, SULFATE ION, Signal transducer and activator of transcription 1-alpha/beta, ... | | Authors: | Pantelejevs, T, Talbot-Cooper, C, Smith, G.L, Hyvonen, M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.0004015 Å) | | Cite: | Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Cell Host Microbe, 30, 2022
|
|
1LR7
 
 | |
1LR9
 
 | |
1LR8
 
 | |
2W8S
 
 | | CRYSTAL STRUCTURE OF A catalytically promiscuous PHOSPHONATE MONOESTER HYDROLASE FROM Burkholderia caryophylli | | Descriptor: | FE (III) ION, GLYCEROL, PHOSPHONATE MONOESTER HYDROLASE, ... | | Authors: | Jonas, S, van Loo, B, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2009-01-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Efficient, Multiply Promiscuous Hydrolase in the Alkaline Phosphatase Superfamily.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4YPV
 
 | | High-resolution structure of a metagenome-derived esterase Est8 | | Descriptor: | Est8 | | Authors: | Pereira, M.R, Maester, C.T, Macedo Lemos, E, Hyvonen, M, Balan, A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | From a metagenomic source to a high-resolution structure of a novel alkaline esterase.
Appl. Microbiol. Biotechnol., 2017
|
|
7OBI
 
 | | Consensus tetratricopeptide repeat protein type RV4 | | Descriptor: | CTPR-rv4, PHOSPHATE ION | | Authors: | Eapen, R.S, Perez-Riba, A, Fischer, G, Itzhaki, L.S, Hyvonen, M. | | Deposit date: | 2021-04-22 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unraveling the Mechanics of a Repeat-Protein Nanospring: From Folding of Individual Repeats to Fluctuations of the Superhelix.
Acs Nano, 16, 2022
|
|
7QE2
 
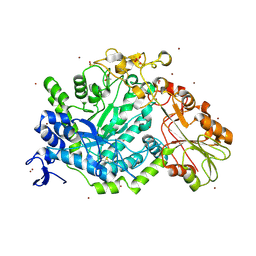 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEE
 
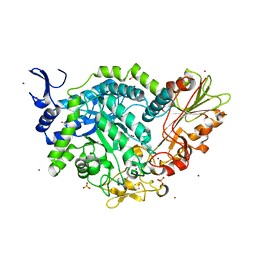 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
5CTP
 
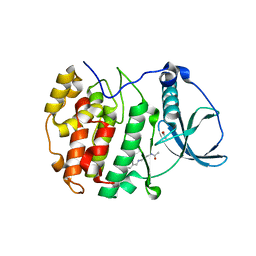 | | Crystal structure of CK2alpha with N-(3-(3-chloro-4-(phenyl)benzylamino)propyl)acetamide bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, N-(3-{[(2-chlorobiphenyl-4-yl)methyl]amino}propyl)acetamide, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5CSH
 
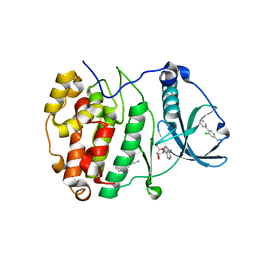 | | Crystal Structure of CK2alpha with Compound 4 bound | | Descriptor: | 1-(2-chlorobiphenyl-4-yl)methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CVF
 
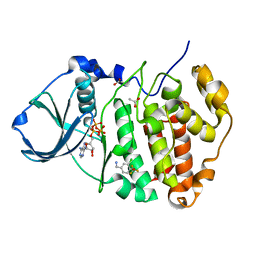 | | Crystal Structure of CK2alpha with Compound 5 bound | | Descriptor: | 1-[3-chloro-4-(trifluoromethoxy)phenyl]methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CU2
 
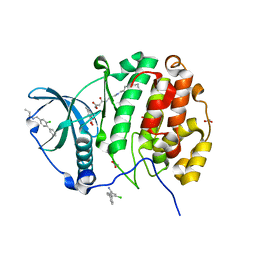 | | Crystal structure of CK2alpha with 2-hydroxy-5-methylbenzoic acid and (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoat bound | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5CVH
 
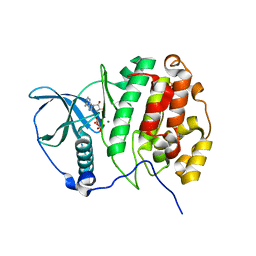 | | Crystal Structure of CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CX9
 
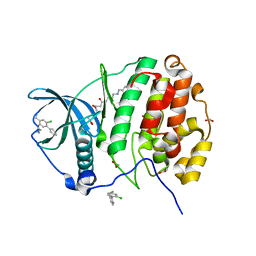 | | Crystal structure of CK2alpha with (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoate bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6HQU
 
 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UPH
 
 | | Crystal Structure of Phosphonate Monoester Hydrolase of Agrobacterium radiobacter | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATASE (SULFURIC ESTER HYDROLASE) PROTEIN | | Authors: | Fischer, G, Loo, B.v, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4UPL
 
 | | Dimeric sulfatase SpAS2 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
