5HGG
 
 | | Crystal structure of uPA in complex with a camelid-derived antibody fragment | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Camelid Derived Antibody Fragment, ... | | Authors: | Yung, K.W.Y, Kromann-Hansen, T, Andreasen, P.A, Ngo, J.C.K. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-01 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
J.Biol.Chem., 291, 2016
|
|
5H7Q
 
 | | Crystal structure of human MNDA PYD domain with MBP tag | | Descriptor: | ACETATE ION, MNDA PYD domain with MBP tag, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5HDO
 
 | |
2NB8
 
 | |
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
2NB7
 
 | |
3D27
 
 | | E. coli methionine aminopeptidase with Fe inhibitor W29 | | Descriptor: | 4-(3-ethylthiophen-2-yl)benzene-1,2-diol, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Chai, S, He, H.Z. | | Deposit date: | 2008-05-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of inhibitors of Escherichia coli methionine aminopeptidase with the Fe(II)-form selectivity and antibacterial activity.
J.Med.Chem., 51, 2008
|
|
8HJA
 
 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | Authors: | Zeng, X, Peng, Y.J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5WQ6
 
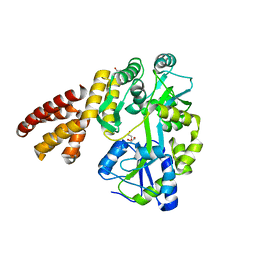 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5WPZ
 
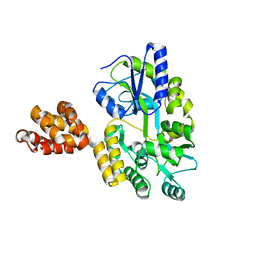 | | Crystal structure of MNDA PYD with MBP tag | | Descriptor: | MBP-hMNDA-PYD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
6A8G
 
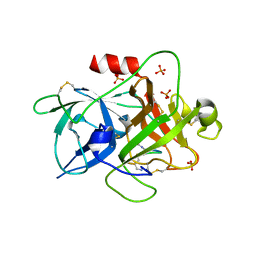 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
6A8N
 
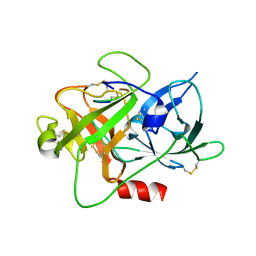 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
4O03
 
 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7VM5
 
 | |
7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | Descriptor: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM6
 
 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM7
 
 | | Crystal structure of inactive uPA in complex with nafamostat | | Descriptor: | (6-carbamimidoylnaphthalen-2-yl) 4-carbamimidamidobenzoate, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
3H0F
 
 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
3H0I
 
 | |
3H0H
 
 | |
3HNB
 
 | |
3HOB
 
 | |
3HNY
 
 | |
4D8D
 
 | |
3QN7
 
 | | Potent and selective bicyclic peptide inhibitor (UK18) of human urokinase-type plasminogen activator(uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, Bicyclic peptide inhibitor, Urokinase-type plasminogen activator | | Authors: | Angelini, A, Cendron, L, Touati, J, Winter, G, Zanotti, G, Heinis, C. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bicyclic peptide inhibitor reveals large contact interface with a protease target
Acs Chem.Biol., 7, 2012
|
|
