2LMS
 
 | | A single GalNAc residue on Threonine-106 modifies the dynamics and the structure of Interferon alpha-2a around the glycosylation site | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Interferon alpha-2 | | Authors: | Ghasriani, H, Belcourt, P.J.F, Sauve, S, Hodgson, D.J, Gingras, G, Brochu, D, Gilbert, M, Aubin, Y. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A single N-acetylgalactosamine residue at threonine 106 modifies the dynamics and structure of interferon alpha2a around the glycosylation site.
J.Biol.Chem., 288, 2013
|
|
2P2V
 
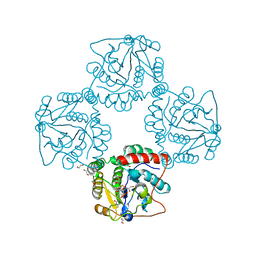 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
2P56
 
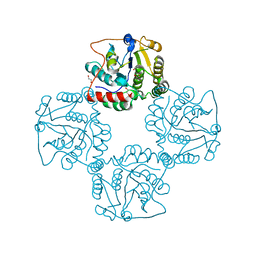 | | Crystal structure of alpha-2,3-sialyltransferase from Campylobacter jejuni in apo form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-14 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
4ZU7
 
 | | X-ray structure if the QdtA 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, double mutant Y17R/R97H, in complex with TDP | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
4ZU4
 
 | | X-ray structure of the 3,4-ketoisomerase domain of FdtD from Shewanella denitrificans | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
4ZU5
 
 | | Crystal structure of the QdtA 3,4-Ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, apo form | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
5U24
 
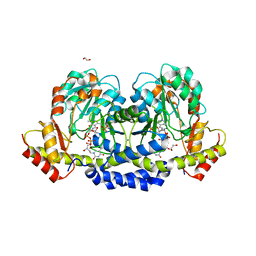 | | X-ray structure of the WlaRG aminotransferase from campylobacter jejuni, K184A mutant in complex with TDP-Fuc3N | | Descriptor: | (2R,3R,4S,5R,6R)-3,5-dihydroxy-4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5TPV
 
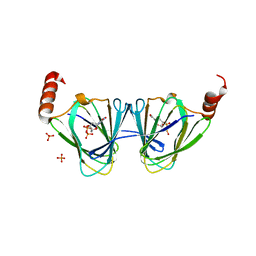 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5TPU
 
 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5U23
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U1Z
 
 | | X-ray structure of the WlarG aminotransferase, apo form, from Campylobacter jejune | | Descriptor: | CHLORIDE ION, Putative aminotransferase, SODIUM ION | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U20
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni, internal PLP-aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U21
 
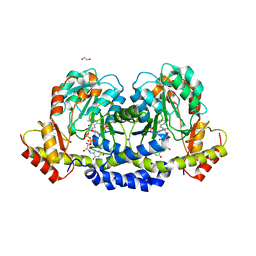 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
1XUU
 
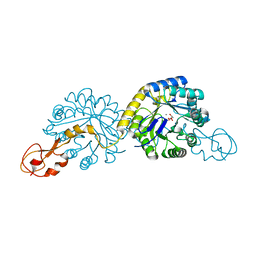 | | Crystal structure of sialic acid synthase (NeuB) in complex with Mn2+ and Malate from Neisseria meningitidis | | Descriptor: | D-MALATE, MANGANESE (II) ION, polysialic acid capsule biosynthesis protein SiaC | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
1XUZ
 
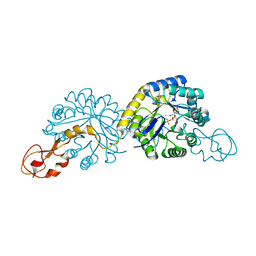 | | Crystal structure analysis of sialic acid synthase (NeuB)from Neisseria meningitidis, bound to Mn2+, Phosphoenolpyruvate, and N-acetyl mannosaminitol | | Descriptor: | 5-DEOXY-5-{[(1S)-1-HYDROXYETHYL]AMINO}-D-GLUCITOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
4LXY
 
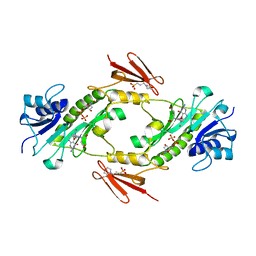 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP and 10-N-Formyl-THF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXT
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp-Qui3N and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LY0
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Glc and 10-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXU
 
 | | dTdp-Fuc3N and 5-N-Formyl-THF | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXQ
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LY3
 
 | | Crystal structure of WlaRD, a sugar 3N-formyl transferase in the presence of dTPD-Qui3N, dTDP-Qui3NFo, and THF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, WlaRD a sugar 3N formyltransferase, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXX
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Fuc3NFo and 5-N-Formyl-THF | | Descriptor: | (2R,3R,4S,5R,6R)-4-(formylamino)-3,5-dihydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | thoden, J.B, goneau, M.-F, gilbert, M, holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
1EYR
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE, CYTIDINE-5'-DIPHOSPHATE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-08 | | Release date: | 2001-02-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
1EZI
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-11 | | Release date: | 2001-02-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
1RO8
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
