7U89
 
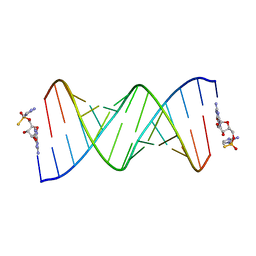 | | Product of 14mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U8A
 
 | | Product of 14mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7LNF
 
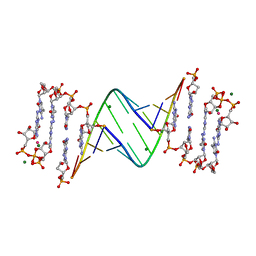 | | 3'-deoxy modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 3'-deoxy-guanosine 5'-monophosphate, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, COBALT (II) ION, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNG
 
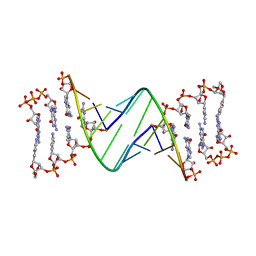 | | TNA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNE
 
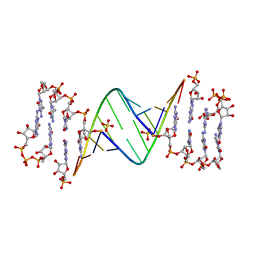 | | ANA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE ARABINOSE-5'-PHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6F52
 
 | |
6F4W
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5I
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | METHANOL, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5A
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F4X
 
 | |
6G7X
 
 | |
5LU0
 
 | | Crystal structure of H. pylori referent strain in complex with PO4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Stefanic, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
5MAL
 
 | |
3EIV
 
 | |
8FEO
 
 | |
8FEP
 
 | |
8FEQ
 
 | |
8FER
 
 | |
8SCJ
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 2h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCL
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 6h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCK
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 4h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCI
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 1h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCG
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 0h (Ground State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 3'-Amino DNA Primer, CALCIUM ION, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCM
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCN
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h (Product State) | | Descriptor: | CALCIUM ION, DIPHOSPHATE, DNA polymerase I, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
