2NQG
 
 | | Calpain 1 proteolytic core inactivated by WR18(S,S), an epoxysuccinyl-type inhibitor. | | Descriptor: | 5-AZANYLIDYNE-N-[(2S)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]-L-NORVALYL-L-ARGINYL-L-TRYPTOPHANAMIDE, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
2MSI
 
 | |
1KXR
 
 | | Crystal Structure of Calcium-Bound Protease Core of Calpain I | | Descriptor: | CALCIUM ION, thiol protease DOMAINS I AND II | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Elce, J.S, Jia, Z, Davies, P.L. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Ca(2+) switch aligns the active site of calpain.
Cell(Cambridge,Mass.), 108, 2002
|
|
1L0S
 
 | |
1L1I
 
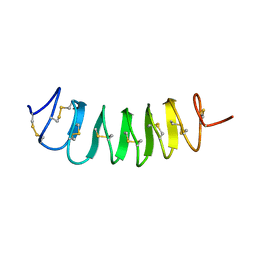 | | Solution Structure of the Tenebrio molitor Antifreeze Protein | | Descriptor: | Thermal hysteresis protein isoform YL-1 (2-14) | | Authors: | Daley, M.E, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-02-16 | | Release date: | 2002-05-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of a beta-helical antifreeze protein.
Biochemistry, 41, 2002
|
|
1M8N
 
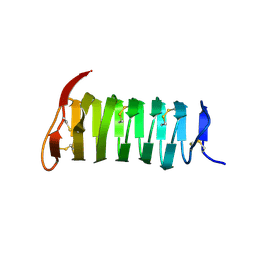 | |
1NP8
 
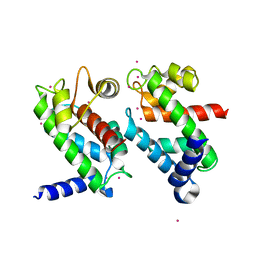 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
2R9C
 
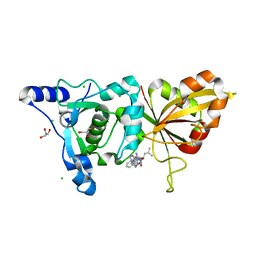 | | Calpain 1 proteolytic core inactivated by ZLAK-3001, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2R9F
 
 | | Calpain 1 proteolytic core inactivated by ZLAK-3002, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
1DVI
 
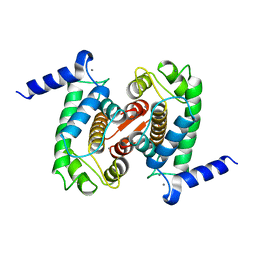 | | CALPAIN DOMAIN VI WITH CALCIUM BOUND | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Cygler, M, Blanchard, H, Grochulski, P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
3RDN
 
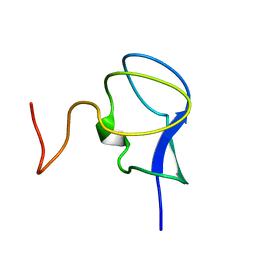 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3NLA
 
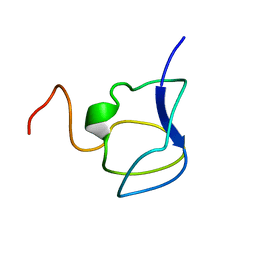 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3DF0
 
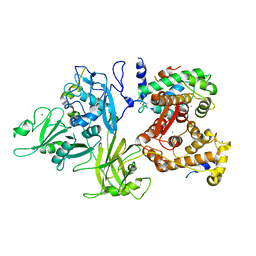 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
1AJ5
 
 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
1C3Z
 
 | | THP12-CARRIER PROTEIN FROM YELLOW MEAL WORM | | Descriptor: | THP12 CARRIER PROTEIN | | Authors: | Soennichsen, F.D. | | Deposit date: | 1999-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands.
Structure Fold.Des., 7, 1999
|
|
1C3Y
 
 | | THP12-CARRIER PROTEIN FROM YELLOW MEAL WORM | | Descriptor: | THP12 CARRIER PROTEIN | | Authors: | Soennichsen, F.D. | | Deposit date: | 1999-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands.
Structure Fold.Des., 7, 1999
|
|
5K8G
 
 | |
4DT5
 
 | | Crystal Structure of Rhagium inquisitor Antifreeze Protein | | Descriptor: | Antifreeze protein, GLYCEROL, SULFATE ION | | Authors: | Meng, W, Nguyen, J.B, Hakim, A, Thakral, D, Zhu, D.F. | | Deposit date: | 2012-02-20 | | Release date: | 2013-03-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of an insect antifreeze protein and its implications for ice binding.
J.Biol.Chem., 288, 2013
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5J6Y
 
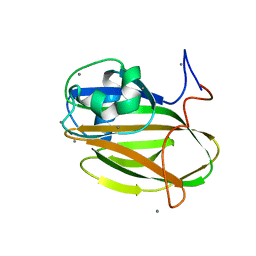 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
1C8A
 
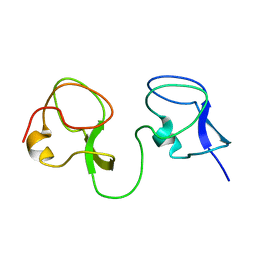 | |
1C89
 
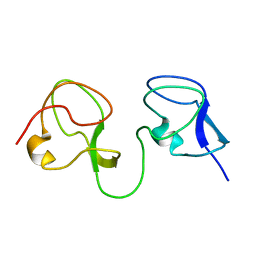 | |
5JUH
 
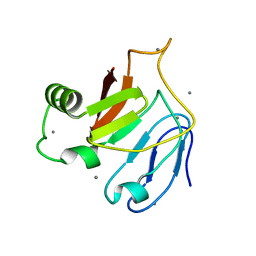 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
1GZI
 
 | | CRYSTAL STRUCTURE OF TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT, AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN | | Authors: | Antson, A.A, Lewis, S, Roper, D.I, Smith, D.J, Hubbard, R.E. | | Deposit date: | 1996-10-25 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Type III Antifreeze Protein from Ocean Pout
To be Published
|
|
