7ML7
 
 | |
4M0U
 
 | | crystal structure of human PRS1 Q133P mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
9BPF
 
 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BOO
 
 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
1IA8
 
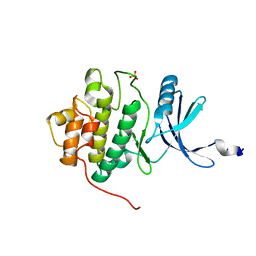 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
6OQ7
 
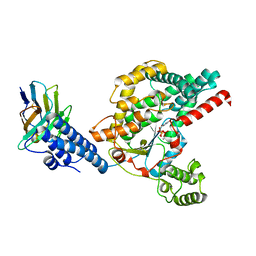 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQ8
 
 | |
6OQ6
 
 | |
7XT3
 
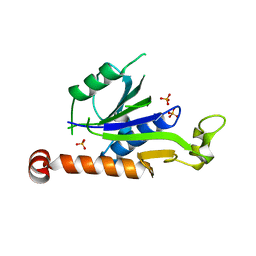 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
5L2I
 
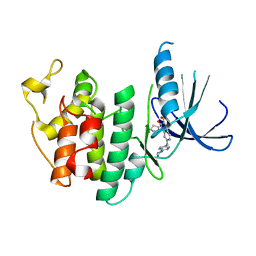 | | The X-ray co-crystal structure of human CDK6 and Palbociclib. | | Descriptor: | 6-ACETYL-8-CYCLOPENTYL-5-METHYL-2-[(5-PIPERAZIN-1-YLPYRIDIN-2-YL)AMINO]PYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, Cyclin-dependent kinase 6 | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-01 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
5L2S
 
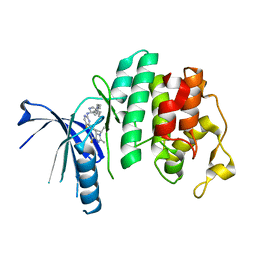 | | The X-ray co-crystal structure of human CDK6 and Abemaciclib. | | Descriptor: | Cyclin-dependent kinase 6, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
5L2W
 
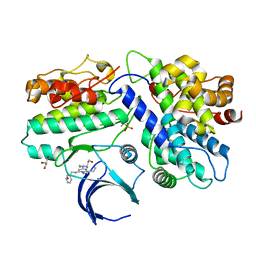 | | The X-ray co-crystal structure of human CDK2/CyclinE and Dinaciclib. | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1, ... | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
5L2T
 
 | | The X-ray co-crystal structure of human CDK6 and Ribociclib. | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(piperazin-1-yl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Cyclin-dependent kinase 6 | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
4TUU
 
 | | Isolated p110a subunit of PI3Ka provides a platform for structure-based drug design | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Deng, Y.-L, Bergqvist, S, Falk, M, Liu, W, Timofeevski, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Protein Sci., 23, 2014
|
|
4TV3
 
 | | Isolated p110a subunit of PI3Ka provides a platform for structure-based drug design | | Descriptor: | 2-amino-8-[trans-4-(2-hydroxyethoxy)cyclohexyl]-6-(6-methoxypyridin-3-yl)-4-methylpyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Deng, Y.-L, Bergqvist, S, Falk, M, Liu, W, Timofeevski, S. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Protein Sci., 23, 2014
|
|
1PI2
 
 | |
1GRC
 
 | |
4LZO
 
 | | Crystal structure of human PRS1 A87T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LYG
 
 | | Crystal structure of human PRS1 E43T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LZN
 
 | | Crystal structure of human PRS1 D65N mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0P
 
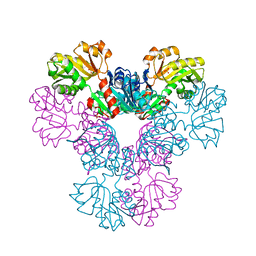 | | Crystal structure of human PRS1 M115T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
7CI1
 
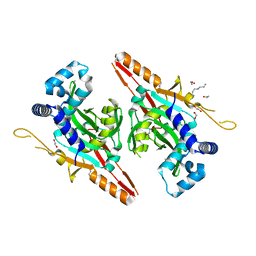 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
7CI2
 
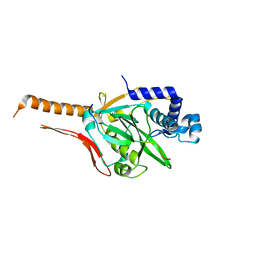 | |
6OQ5
 
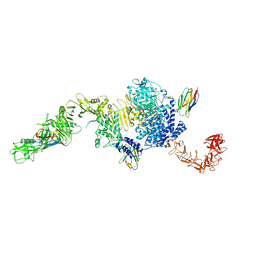 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6C0B
 
 | | Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-2, MALONATE ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2017-12-28 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of frizzled proteins byClostridium difficiletoxin B.
Science, 360, 2018
|
|
