2CIU
 
 | |
2YH3
 
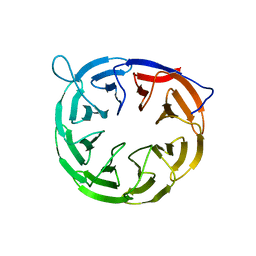 | | The structure of BamB from E. coli | | Descriptor: | LIPOPROTEIN YFGL | | Authors: | Albrecht, R, Zeth, K. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
5A1Q
 
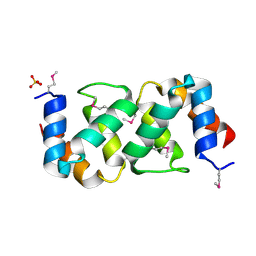 | |
5FZQ
 
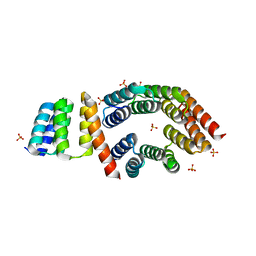 | | Designed TPR Protein M4N | | Descriptor: | DESIGNED TPR PROTEIN, SULFATE ION | | Authors: | Albrecht, R, Zhu, H, Hartmann, M.D. | | Deposit date: | 2016-03-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Origin of a folded repeat protein from an intrinsically disordered ancestor.
Elife, 5, 2016
|
|
5FZR
 
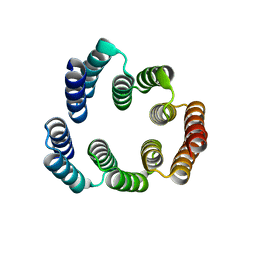 | |
5FZS
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
2YHC
 
 | | Structure of BamD from E. coli | | Descriptor: | UPF0169 LIPOPROTEIN YFIO, UREA | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
2YH9
 
 | |
2YH6
 
 | |
2YMS
 
 | |
4C4V
 
 | |
2YH5
 
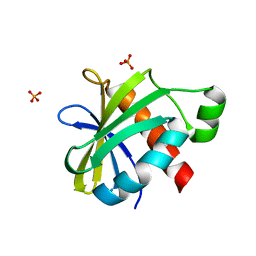 | | Structure of the C-terminal domain of BamC | | Descriptor: | DAPX PROTEIN, PHOSPHATE ION | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
1J5A
 
 | | STRUCTURAL BASIS FOR THE INTERACTION OF ANTIBIOTICS WITH THE PEPTIDYL TRANSFERASE CENTER IN EUBACTERIA | | Descriptor: | 23S RRNA, CLARITHROMYCIN, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1Y69
 
 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | Deposit date: | 2004-12-04 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
2D3O
 
 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
5OH8
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Rolipram | | Descriptor: | Cereblon isoform 4, ROLIPRAM, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH4
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH2
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OHB
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidin-2-one (Valerolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH1
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Aminoglutethimide | | Descriptor: | (3~{R})-3-(4-aminophenyl)-3-ethyl-piperidine-2,6-dione, Cereblon isoform 4, S-Thalidomide, ... | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH9
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thiazolidine-2,4-dione | | Descriptor: | 1,3-thiazole-2,4-dione, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH3
 
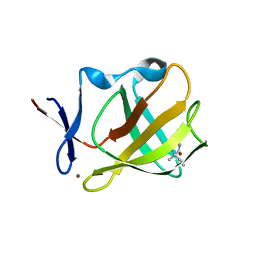 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Ethosuximide | | Descriptor: | (3~{S})-3-ethyl-3-methyl-pyrrolidine-2,5-dione, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OHA
 
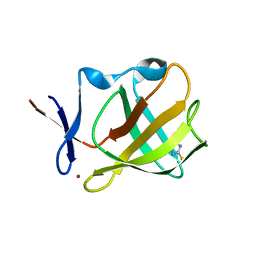 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with 2-Thiohydantoin | | Descriptor: | 2-sulfanylideneimidazol-4-one, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH7
 
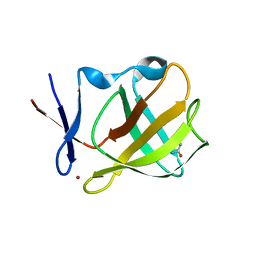 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Imidazolidine-2,4-dione (Hydantoin) | | Descriptor: | Cereblon isoform 4, ZINC ION, imidazolidine-2,4-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
