6B0X
 
 | |
6B23
 
 | |
6B3E
 
 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
2G5N
 
 | |
2GBJ
 
 | |
2GBN
 
 | |
2GBK
 
 | |
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
3CB0
 
 | | CobR | | Descriptor: | 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE, FLAVIN MONONUCLEOTIDE | | Authors: | Lawrence, A.D, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis
J.Biol.Chem., 283, 2008
|
|
3D0X
 
 | |
6BIC
 
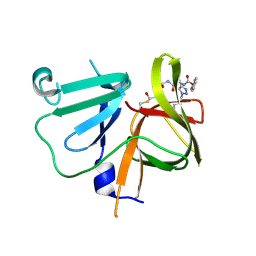 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BIB
 
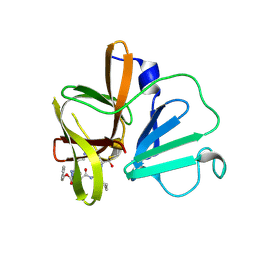 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
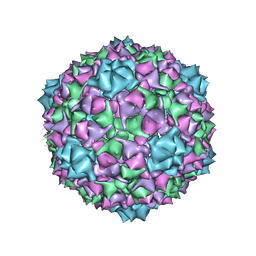
6C22
 
 | |
2GZM
 
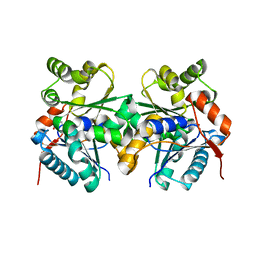 | | Crystal Structure of the Glutamate Racemase from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | May, M, Santarsiero, B.D, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design.
J.Mol.Biol., 371, 2007
|
|
3D0U
 
 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BSK
 
 | | Human PIM1 kinase in complex with compound 12b | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-[6-(propan-2-ylamino)-1H-indazol-1-yl]pyrazin-2-yl}benzoic acid, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-12-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazine derivatives as inhibitors of CK2 and PIM kinases.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6BI8
 
 | |
6C21
 
 | |
6BWK
 
 | |
2ORM
 
 | | Crystal Structure of the 4-Oxalocrotonate Tautomerase Homologue DmpI from Helicobacter pylori. | | Descriptor: | Probable tautomerase HP0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Czerwinski, R.M, Kern, A.D. | | Deposit date: | 2007-02-03 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
2OPY
 
 | | Smac mimic bound to BIR3-XIAP | | Descriptor: | 1-({2-[(1S)-1-AMINOETHYL]-1,3-OXAZOL-4-YL}CARBONYL)-L-PROLYL-L-TRYPTOPHAN, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Wist, A.D, Lu, G, Riedl, S.J, Shi, Y, McLendon, G.L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
3DMT
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetate inhibitor | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Guido, R.V.C, Balliano, T.L, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Crystallographic Studies on Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in Complex with Iodoacetate.
Letters in drug design & discovery, 6, 2009
|
|
2OPZ
 
 | | AVPF bound to BIR3-XIAP | | Descriptor: | AVPF (Smac homologue, N-terminal tetrapeptide), Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Wist, A.D. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
2P4V
 
 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
