2QX8
 
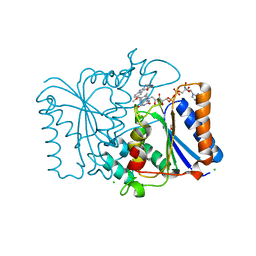 | | Crystal Structure of Quinone Reductase II | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
4JX6
 
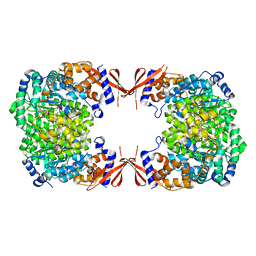 | |
6DLV
 
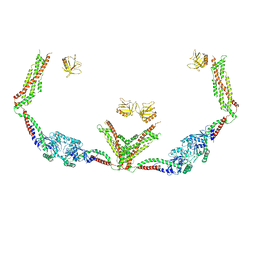 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
5T1H
 
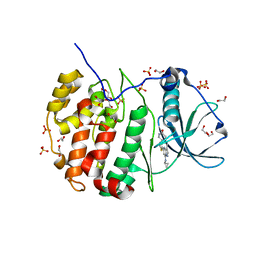 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[3-(6-oxo-1,6-dihydropyridin-3-yl)thiophen-2-yl]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Feguson, A.D. | | Deposit date: | 2016-08-19 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
6TFL
 
 | | Lsm protein (SmAP) from Halobacterium salinarum | | Descriptor: | CALCIUM ION, GLYCEROL, RNA-binding protein Lsm, ... | | Authors: | Nikulin, A.D, Fando, M.S, Lekontseva, N.V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure and RNA-Binding Properties of Lsm Protein from Halobacterium salinarum.
Biochemistry Mosc., 86, 2021
|
|
5T6F
 
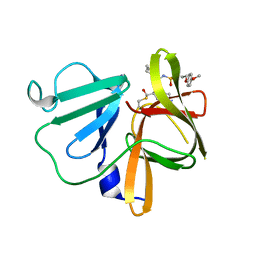 | | 1.90 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (orthorhombic P form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5T6D
 
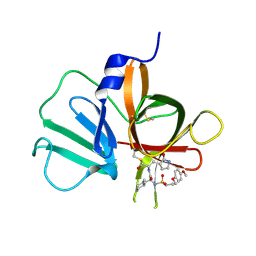 | | 2.10 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5T6G
 
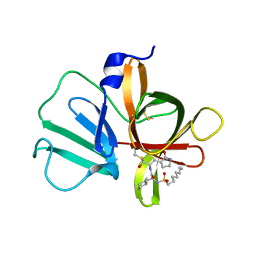 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
6TIW
 
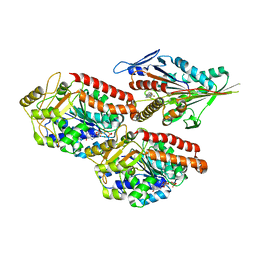 | | Human kinesin-5 motor domain in the GSK state bound to microtubules (Conformation 2) | | Descriptor: | 6-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-quinolin-2-one, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-11-22 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
6TA4
 
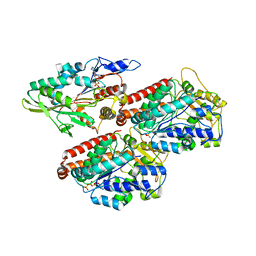 | | Human kinesin-5 motor domain in the AMPPNP state bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A.P, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
6MDR
 
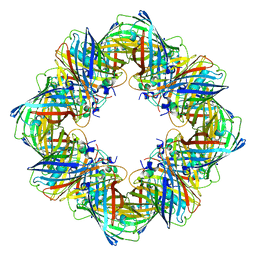 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
6T8S
 
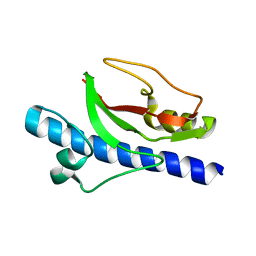 | |
6TA3
 
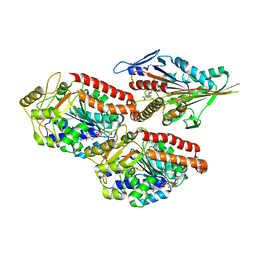 | | Human kinesin-5 motor domain in the GSK-1 state bound to microtubules (Conformation 1) | | Descriptor: | 6-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-quinolin-2-one, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
6E99
 
 | |
2V7A
 
 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
6E6N
 
 | | Pheromone from Euplotes raikovi, Er-13 | | Descriptor: | Pheromone from Euplotes raikovi Er-13 | | Authors: | Finke, A.D, Marsh, M.E. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (1.363 Å) | | Cite: | Ab initio crystal structure determination of Euplotes raikovi pheromones from high-resolution data
To Be Published
|
|
6E6O
 
 | | Pheromone from Euplotes raikovi, Er-1 | | Descriptor: | Mating pheromone Er-1/Er-3 | | Authors: | Finke, A.D, Marsh, M.E. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Ab initio crystal structure determination of Euplotes raikovi pheromones from high-resolution data
To Be Published
|
|
6E9W
 
 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
2VH0
 
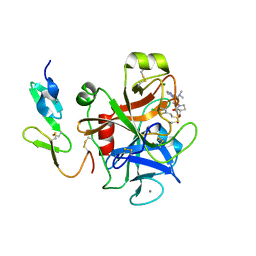 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-25 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
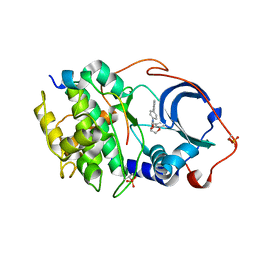
6E9L
 
 | |
4L1P
 
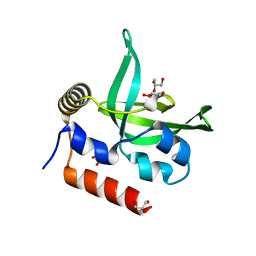 | |
6MUZ
 
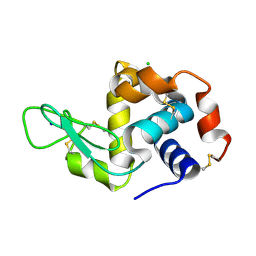 | | Lysozyme, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Kriksunov, I, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6MSF
 
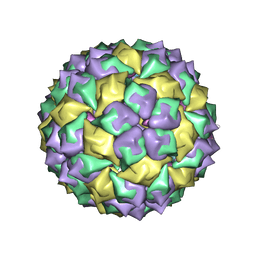 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | Authors: | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
6MUY
 
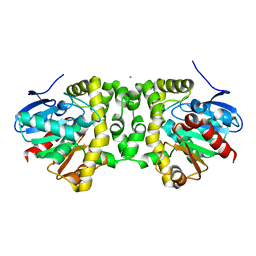 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6ED6
 
 | |
