3BJZ
 
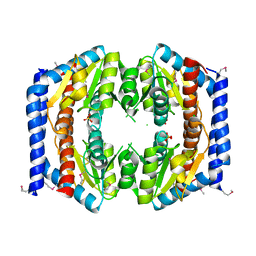 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
3ESQ
 
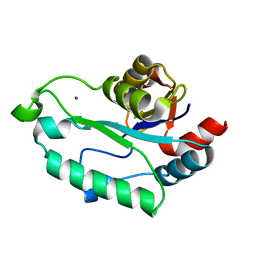 | |
3ESR
 
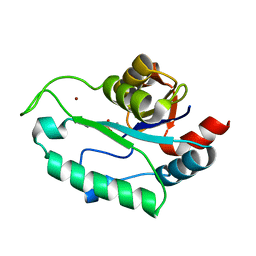 | | Crystal Structure of D,D-heptose1.7-bisphosphate phosphatase from E. coli in complex with calcium and phosphate | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, PHOSPHATE ION, ... | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli.
To be Published
|
|
2I22
 
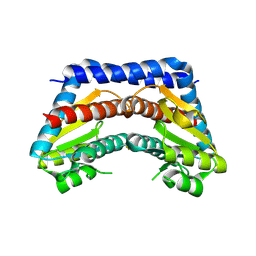 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | Authors: | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2W
 
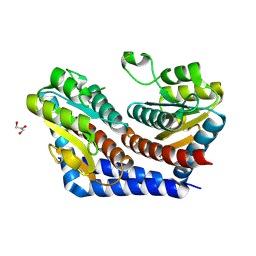 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2OVB
 
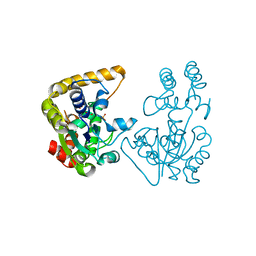 | | Crystal Structure of StaL-sulfate complex | | Descriptor: | SULFATE ION, StaL | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis.
J.Biol.Chem., 282, 2007
|
|
2OV8
 
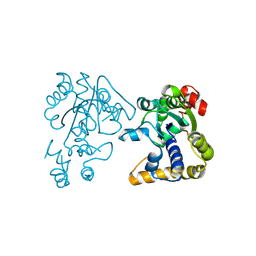 | |
2OVF
 
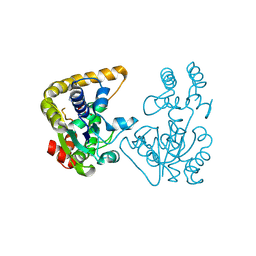 | | Crystal Structure of StaL-PAP complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, StaL | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis.
J.Biol.Chem., 282, 2007
|
|
5FBT
 
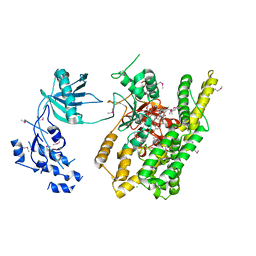 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FBS
 
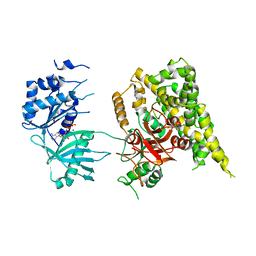 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with ADP and magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoenolpyruvate synthase | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FGZ
 
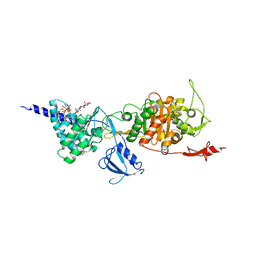 | | E. coli PBP1b in complex with FPI-1465 | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
2FQ6
 
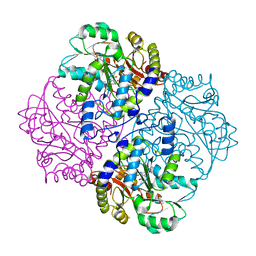 | |
2GQN
 
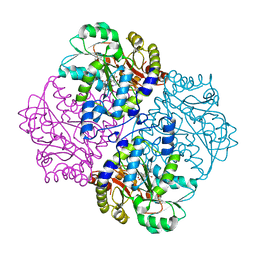 | |
5UXD
 
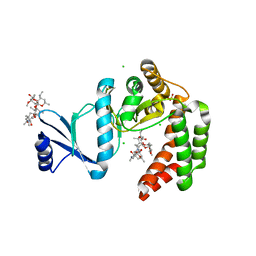 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXA
 
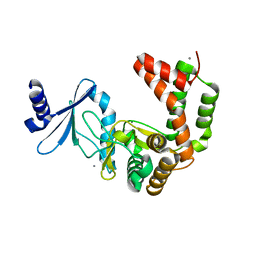 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXC
 
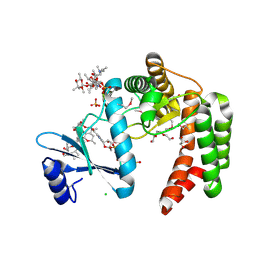 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-16 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5US1
 
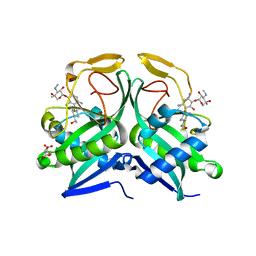 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5UXB
 
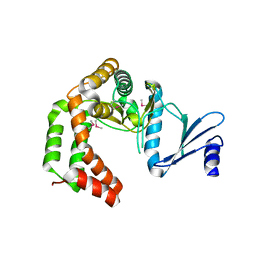 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
1X92
 
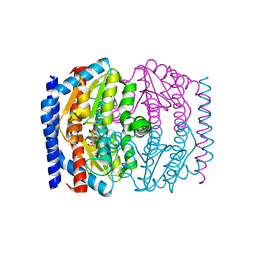 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA PHOSPHOHEPTOSE ISOMERASE IN COMPLEX WITH REACTION PRODUCT D-GLYCERO-D-MANNOPYRANOSE-7-PHOSPHATE | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, PHOSPHOHEPTOSE ISOMERASE | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants.
J.Biol.Chem., 283, 2008
|
|
6CFD
 
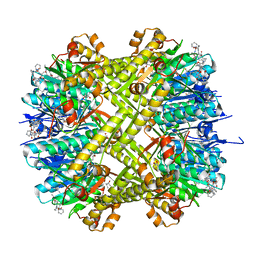 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
1J7U
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Complex | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7L
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7I
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
