2Q0U
 
 | | Structure of Pectenotoxin-2 and Latrunculin B Bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Allingham, J.S, Miles, C.O, Rayment, I. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural basis for regulation of actin polymerization by pectenotoxins.
J.Mol.Biol., 371, 2007
|
|
4FUQ
 
 | | Crystal structure of apo MatB from Rhodopseudomonas palustris | | Descriptor: | GLYCEROL, Malonyl CoA synthetase, SULFATE ION | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-06-28 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Guided Expansion of the Substrate Range of Methylmalonyl Coenzyme A Synthetase (MatB) of Rhodopseudomonas palustris.
Appl.Environ.Microbiol., 78, 2012
|
|
4FUT
 
 | | Crystal structure of ATP bound MatB from Rhodopseudomonas palustris | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-06-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Expansion of the Substrate Range of Methylmalonyl Coenzyme A Synthetase (MatB) of Rhodopseudomonas palustris.
Appl.Environ.Microbiol., 78, 2012
|
|
4HDM
 
 | | Crystal Structure of ArsAB in Complex with p-cresol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDS
 
 | | Crystal Structure of ArsAB in Complex with Phenol. | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4GXQ
 
 | | Crystal Structure of ATP bound RpMatB-BxBclM chimera B1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
4GXR
 
 | | Structure of ATP bound RpMatB-BxBclM chimera B3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, GLYCEROL, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
4HDR
 
 | | Crystal Structure of ArsAB in Complex with 5,6-dimethylbenzimidazole | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, ArsA, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDK
 
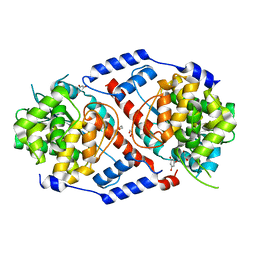 | | Crystal Structure of ArsAB in Complex with Phloroglucinol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDN
 
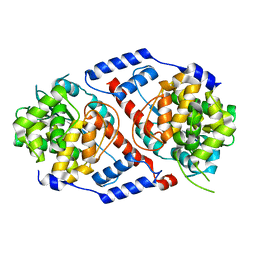 | | Crystal Structure of ArsAB in the Substrate-Free State. | | Descriptor: | ArsA, ArsB | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HUT
 
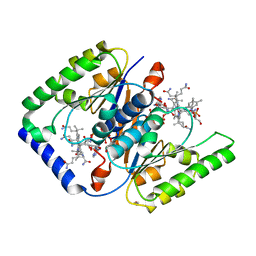 | | Structure of ATP:co(I)rrinoid adenosyltransferase (CobA) from Salmonella enterica in complex with four and five-coordinate cob(II)alamin and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, ... | | Authors: | Moore, T.C, Newmister, S.A, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2012-11-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of Four-Coordinate Cob(II)alamin Formation in the Active Site of the Salmonella enterica ATP:Co(I)rrinoid Adenosyltransferase Enzyme: Critical Role of Residues Phe91 and Trp93.
Biochemistry, 51, 2012
|
|
4IFF
 
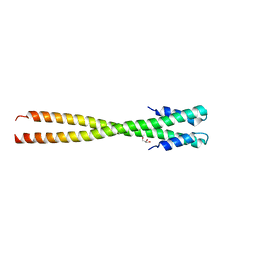 | | Structural organization of FtsB, a transmembrane protein of the bacterial divisome | | Descriptor: | Fusion of phage phi29 Gp7 protein and Cell division protein FtsB, GLYCEROL | | Authors: | LaPointe, L.M, Taylor, K.C, Subramaniam, S, Khadria, A, Rayment, I, Senes, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Organization of FtsB, a Transmembrane Protein of the Bacterial Divisome.
Biochemistry, 52, 2013
|
|
1HXP
 
 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
1ECQ
 
 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO 4-DEOXYGLUCARATE | | Descriptor: | 4-DEOXYGLUCARATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EC8
 
 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO PRODUCT 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EC7
 
 | | E. COLI GLUCARATE DEHYDRATASE NATIVE ENZYME | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1JDF
 
 | | Glucarate Dehydratase from E.coli N341D mutant | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JCT
 
 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1EBH
 
 | |
1EC9
 
 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1MUS
 
 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
1MUH
 
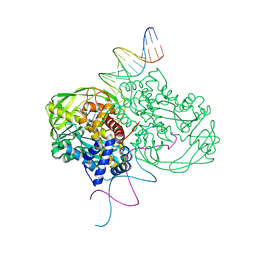 | | CRYSTAL STRUCTURE OF TN5 TRANSPOSASE COMPLEXED WITH TRANSPOSON END DNA | | Descriptor: | DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Davies, D.R, Goryshin, I.Y, Reznikoff, W.S, Rayment, I. | | Deposit date: | 2002-09-23 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the Tn5 synaptic complex transposition intermediate.
Science, 289, 2000
|
|
1P90
 
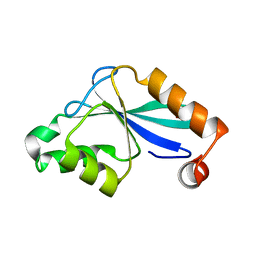 | | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 resolution | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein | | Authors: | Dyer, D.H, Rubio, L.M, Thoden, J.B, Holden, H.M, Ludden, P.W, Rayment, I. | | Deposit date: | 2003-05-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 A resolution
J.Biol.Chem., 278, 2003
|
|
1PZ4
 
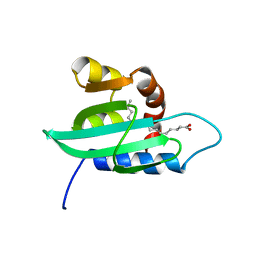 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | Descriptor: | PALMITIC ACID, sterol carrier protein 2 | | Authors: | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
1MM8
 
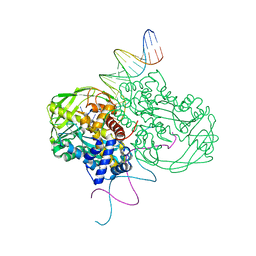 | | Crystal structure of Tn5 Transposase complexed with ME DNA | | Descriptor: | MANGANESE (II) ION, ME DNA non-transferred strand, ME DNA transferred strand, ... | | Authors: | Steiniger-White, M, Bhasin, A, Lovell, S, Rayment, I, Reznikoff, W.S. | | Deposit date: | 2002-09-03 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for "unseen" Transposase--DNA contacts
J.Mol.Biol., 322, 2002
|
|
