4YED
 
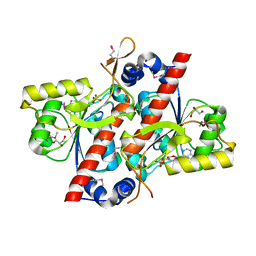 | | TcdA (CsdL) | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Kim, S, Park, S.Y. | | Deposit date: | 2015-02-24 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity
J.Mol.Biol., 427, 2015
|
|
4PRL
 
 | |
4PRK
 
 | |
7UIY
 
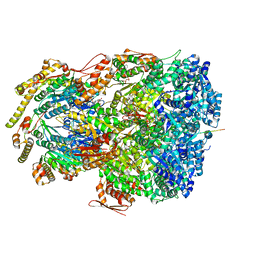 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6KD7
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of geranylgeranyl pyrophosphate synthase (crtE) from Nonlabens dokdonensis DSW-6.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
7VID
 
 | |
1JE4
 
 | |
1RYU
 
 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
5Y9D
 
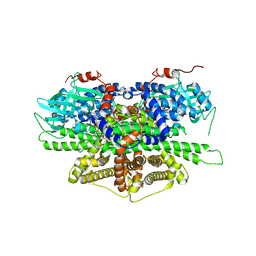 | | Crystal structure of acyl-coA oxidase1 from Yarrowia lipolytica | | Descriptor: | Acyl-coenzyme A oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2017-08-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the substrate specificity of acyl-CoA oxidase1 from Yarrowia lipolytica for short-chain dicarboxylyl-CoAs.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YS9
 
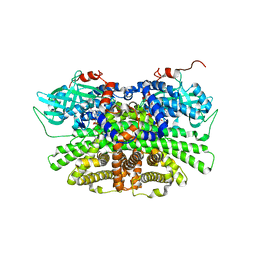 | | Crystal structure of acyl-coA oxidase3 from Yarrowia lipolytica | | Descriptor: | Acyl-coenzyme A oxidase 3, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Acyl-CoA Oxidase 3 fromYarrowia lipolyticawith Specificity for Short-Chain Acyl-CoA.
J. Microbiol. Biotechnol., 28, 2018
|
|
4LW4
 
 | |
4LW2
 
 | |
6L33
 
 | |
3ACP
 
 | | Crystal Structure of Yeast Rpn14, a Chaperone of the 19S Regulatory Particle of the Proteasome | | Descriptor: | WD repeat-containing protein YGL004C | | Authors: | Kim, S, Saeki, Y, Suzuki, A, Takagi, K, Fukunaga, K, Yamane, T, Kato, K, Tanaka, K, Mizushima, T. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Rpn14, a chaperone of the 19S regulatory particle of the proteasome
J.Biol.Chem., 285, 2010
|
|
1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
3OTP
 
 | |
2QCZ
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
2QDF
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | MAGNESIUM ION, Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
3VL1
 
 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7Y1H
 
 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(5-chloranyl-4-methyl-benzimidazol-1-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
