1HRA
 
 | | THE SOLUTION STRUCTURE OF THE HUMAN RETINOIC ACID RECEPTOR-BETA DNA-BINDING DOMAIN | | Descriptor: | RETINOIC ACID RECEPTOR, ZINC ION | | Authors: | Knegtel, R.M.A, Katahira, M, Schilthuis, J.G, Bonvin, A.M.J.J, Boelens, R, Eib, D, Van Der Saag, P.T, Kaptein, R. | | Deposit date: | 1993-07-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the human retinoic acid receptor-beta DNA-binding domain.
J.Biomol.NMR, 3, 1993
|
|
1LCC
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1CCN
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Direct NOE refinement of biomolecular structures using 2D NMR data
J.Biomol.NMR, 1, 1991
|
|
1CCM
 
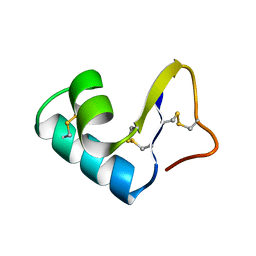 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
1CJG
 
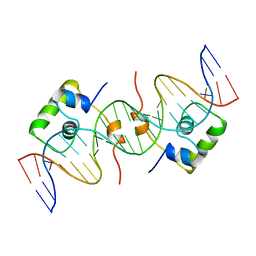 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1HUE
 
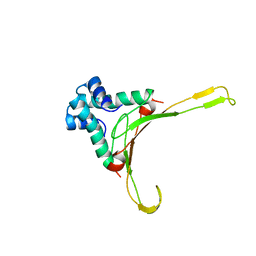 | | HISTONE-LIKE PROTEIN | | Descriptor: | HU PROTEIN | | Authors: | Vis, H, Mariani, M, Vorgias, C.E, Wilson, K.S, Kaptein, R, Boelens, R. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HU protein from Bacillus stearothermophilus.
J.Mol.Biol., 254, 1995
|
|
1SFV
 
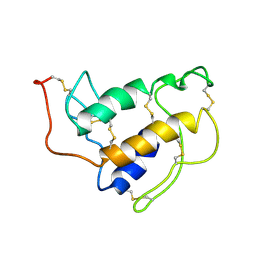 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1SFW
 
 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, 18 STRUCTURES | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-23 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1LQC
 
 | | LAC REPRESSOR HEADPIECE (RESIDUES 1-56), NMR, 32 STRUCTURES | | Descriptor: | LAC REPRESSOR | | Authors: | Slijper, M, Bonvin, A.M.J.J, Boelens, R, Kaptein, R. | | Deposit date: | 1996-08-13 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator.
J.Mol.Biol., 259, 1996
|
|
1RGD
 
 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1ARR
 
 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
1ARQ
 
 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
2BJC
 
 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | Authors: | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
1KFT
 
 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
3PHY
 
 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Dux, P, Rubinstenn, G, Vuister, G.W, Boelens, R, Mulder, F.A.A, Hard, K, Hoff, W.D, Kroon, A, Crielaard, W, Hellingwerf, K.J, Kaptein, R. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the photoactive yellow protein.
Biochemistry, 37, 1998
|
|
1E0E
 
 | | N-terminal zinc-binding HHCC domain of HIV-2 integrase | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 INTEGRASE, ZINC ION | | Authors: | Eijkelenboom, A.P.A.M, Van Den ent, F.M.I, Plasterk, R.H.A, Kaptein, R, Boelens, R. | | Deposit date: | 2000-03-25 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the Dimeric N-Terminal Hhcc Domain of HIV-2 Integrase
J.Biomol.NMR, 18, 2000
|
|
1WCO
 
 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
1MNT
 
 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
1T3O
 
 | | Solution structure of CsrA, a bacterial carbon storage regulatory protein | | Descriptor: | Carbon storage regulator | | Authors: | Koharudin, L.M.I, Georgiou, T, Kleanthous, C, Geoffrey, R, Kaptein, R, Boelens, R. | | Deposit date: | 2004-04-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A model for RNA binding by the bacterial carbon storage regulatory protein, CsrA
To be Published
|
|
1QEY
 
 | | NMR Structure Determination of the Tetramerization Domain of the MNT Repressor: An Asymmetric A-Helical Assembly in Slow Exchange | | Descriptor: | PROTEIN (REGULATORY PROTEIN MNT) | | Authors: | Nooren, I.M.A, George, A.V.E, Kaptein, R, Sauer, R.T, Boelens, R. | | Deposit date: | 1999-04-03 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils.
Nat.Struct.Biol., 6, 1999
|
|
2VKC
 
 | | Solution structure of the B3BP Smr domain | | Descriptor: | NEDD4-BINDING PROTEIN 2 | | Authors: | Diercks, T, Ab, E, Daniels, M.A, deJong, R.N, Besseling, R, Kaptein, R, Folkers, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the DNA- Binding Activity of the B3BP-Smr Domain.
J.Mol.Biol., 383, 2008
|
|
1XFQ
 
 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1XFN
 
 | | NMR structure of the ground state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1Z00
 
 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
