1IB8
 
 | | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE | | Descriptor: | CONSERVED PROTEIN SP14.3 | | Authors: | Yu, L, Gunasekera, A.H, Mack, J, Olejniczak, E.T, Chovan, L.E, Ruan, X, Towne, D.L, Lerner, C.G, Fesik, S.W. | | Deposit date: | 2001-03-27 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE
J.Mol.Biol., 311, 2001
|
|
1NNY
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | Descriptor: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1NO6
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | Descriptor: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1Q59
 
 | | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2 | | Descriptor: | Early antigen protein R | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2
J.Mol.Biol., 332, 2003
|
|
1K3K
 
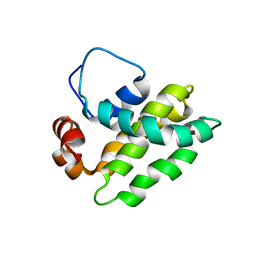 | | Solution Structure of a Bcl-2 Homolog from Kaposi's Sarcoma Virus | | Descriptor: | functional anti-apoptotic factor vBCL-2 homolog | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5WFQ
 
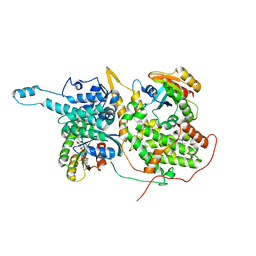 | | Ligand-bound Ras:SOS:Ras complex | | Descriptor: | 7-chloranyl-~{N}-(3-chloranyl-4-fluoranyl-phenyl)-1,2,3,4-tetrahydroacridin-9-amine, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Sun, Q, Phan, J, Burns, M.C, Fesik, S.W. | | Deposit date: | 2017-07-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling.
Anal. Biochem., 548, 2018
|
|
4IPC
 
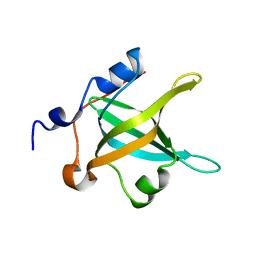 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
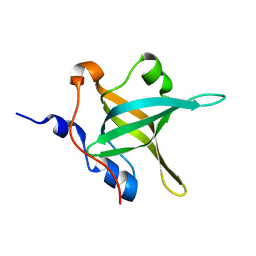 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
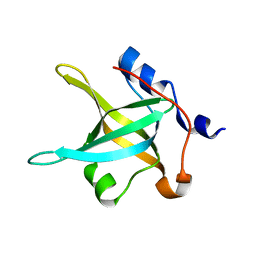 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
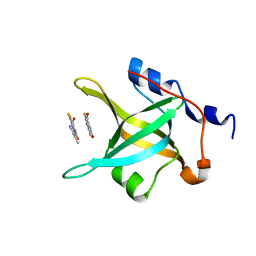 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
5WFR
 
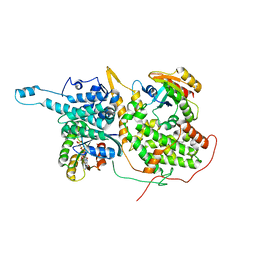 | | Ligand-bound Ras:SOS:Ras complex | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Sun, Q, Phan, J, Burns, M.C, Fesik, S.W. | | Deposit date: | 2017-07-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling.
Anal. Biochem., 548, 2018
|
|
1A1Z
 
 | | FADD DEATH EFFECTOR DOMAIN, F25G MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
1A1W
 
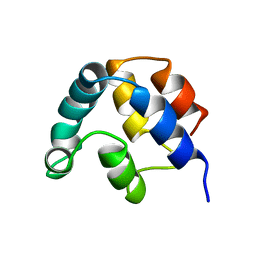 | | FADD DEATH EFFECTOR DOMAIN, F25Y MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
1BC9
 
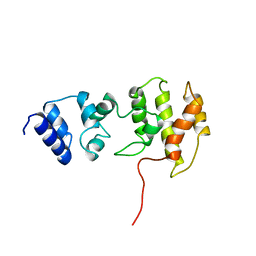 | | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOHESIN-1 | | Authors: | Betz, S.F, Schnuchel, A, Wang, H, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1998-05-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytohesin-1 (B2-1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1C9Q
 
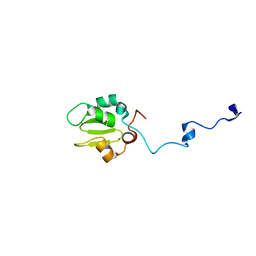 | |
1BXL
 
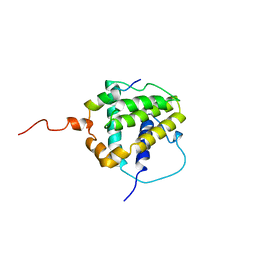 | | STRUCTURE OF BCL-XL/BAK PEPTIDE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAK PEPTIDE, BCL-XL | | Authors: | Sattler, M, Liang, H, Nettesheim, D, Meadows, R.P, Harlan, J.E, Eberstadt, M, Yoon, H, Shuker, S.B, Chang, B.S, Minn, A.J, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1996-10-16 | | Release date: | 1997-10-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis.
Science, 275, 1997
|
|
4LUV
 
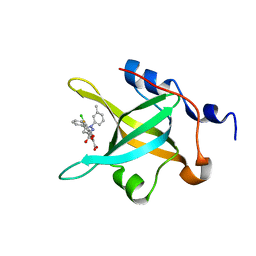 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LUO
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
1DHM
 
 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
4LW1
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LWC
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-[3-chloro-4-({4-[1-(3,4-dichlorophenyl)-1H-pyrazol-5-yl]benzyl}carbamothioyl)phenyl]furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LUZ
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4NB3
 
 | | Crystal structure of RPA70N in complex with a 3,4 dichlorophenylalanine ATRIP derived peptide | | Descriptor: | 3,4 dichlorophenylalanine ATRIP derived peptide, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Souza-Fagundes, E.M, Luzwik, J.W, Cortez, D, Olejniczak, O.T, Waterson, A.G, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
J.Med.Chem., 57, 2014
|
|
6BVJ
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 5-chloro-N-{1-[(5-chloro-1H-indol-3-yl)methyl]piperidin-4-yl}-L-tryptophanamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Abbott, J, Fesik, S.W. | | Deposit date: | 2017-12-13 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Discovery of Aminopiperidine Indoles That Activate the Guanine Nucleotide Exchange Factor SOS1 and Modulate RAS Signaling.
J. Med. Chem., 61, 2018
|
|
4NYM
 
 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-[1-(1H-indol-3-ylmethyl)piperidin-4-yl]-L-tryptophanamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5529 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
