1EH4
 
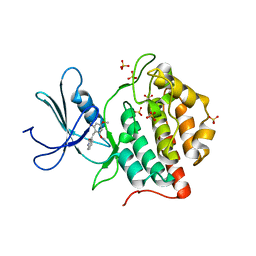 | | BINARY COMPLEX OF CASEIN KINASE-1 FROM S. POMBE WITH AN ATP COMPETITIVE INHIBITOR, IC261 | | Descriptor: | 3-[(2,4,6-TRIMETHOXY-PHENYL)-METHYLENE]-INDOLIN-2-ONE, CASEIN KINASE-1, SULFATE ION | | Authors: | Mashhoon, N, Demaggio, A.J, Tereshko, V, Bergmeier, S.C, Egli, M, Hoekstra, M.F, Kuret, J. | | Deposit date: | 2000-02-18 | | Release date: | 2001-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Conformation-Selective Casein Kinase-1 Inhibitor
J.Biol.Chem., 275, 2000
|
|
2MXP
 
 | |
8DEA
 
 | |
8DG6
 
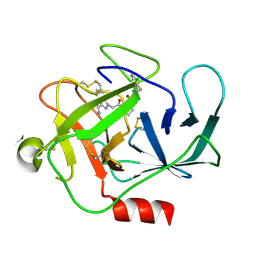 | |
8D95
 
 | |
2BZG
 
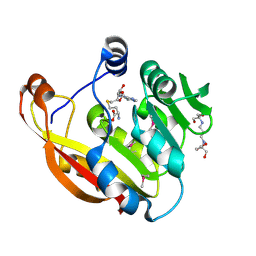 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
5EP6
 
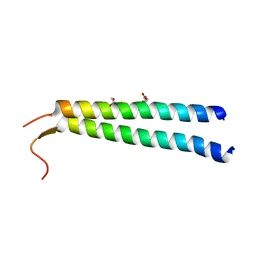 | | The crystal structure of NAP1 in complex with TBK1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EOF
 
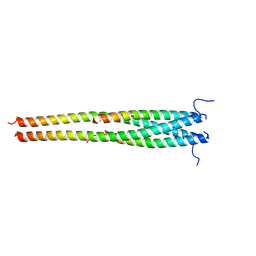 | | Crystal structure of OPTN NTD and TBK1 CTD complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
4BHM
 
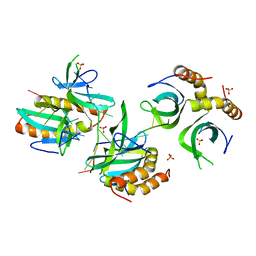 | | The crystal structure of MoSub1-DNA complex reveals a novel DNA binding mode | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP)-3', MOSUB1 TRANSCRIPTION COFACTOR, SULFATE ION | | Authors: | Huang, J, Liu, H, Zhao, Y, Huang, D, liu, J, Peng, Y. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-23 | | Last modified: | 2015-03-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
5EOA
 
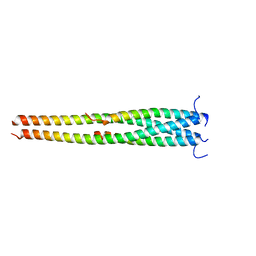 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
6U37
 
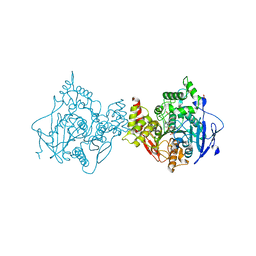 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U3P
 
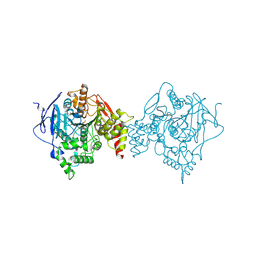 | | Binary complex of native hAChE with oxime reactivator LG-703 | | Descriptor: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U34
 
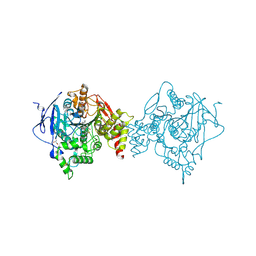 | | Binary complex of native hAChE with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
4OC8
 
 | | DNA modification-dependent restriction endonuclease AspBHI | | Descriptor: | PHOSPHATE ION, restriction endonuclease AspBHI | | Authors: | Horton, J.R. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI.
Sci Rep, 4, 2014
|
|
8DT4
 
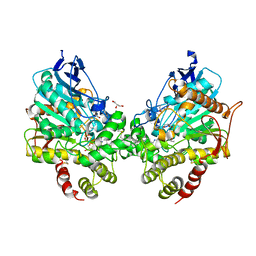 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime MMB4 (POX-hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT2
 
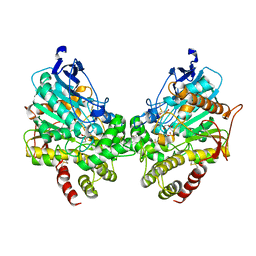 | | X-ray structure of human acetylcholinesterase inhibited by paraoxon (POX-hAChE) | | Descriptor: | Acetylcholinesterase, DIETHYL PHOSPHONATE, DIMETHYL SULFOXIDE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT7
 
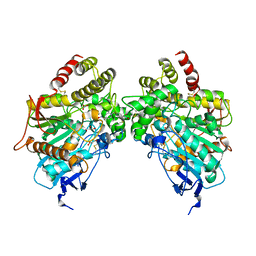 | | X-ray structure of human acetylcholinesterase in complex with oxime MMB4 (hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT5
 
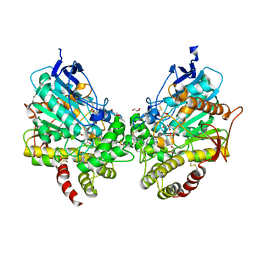 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime RS170B (POX-hAChE-RS170B) | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DKB
 
 | |
7YVN
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH281 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVP
 
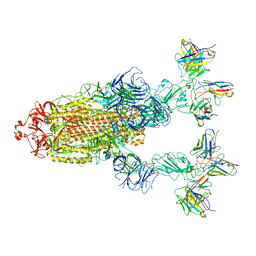 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272/281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVG
 
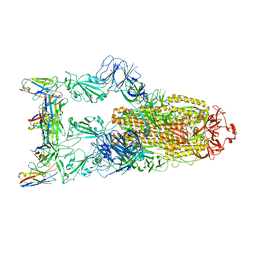 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVK
 
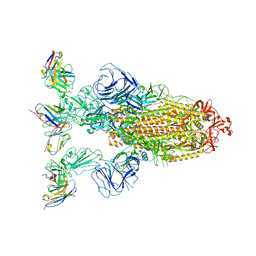 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVO
 
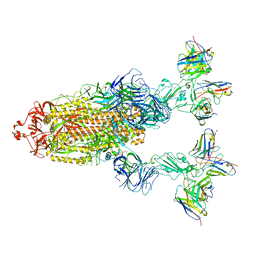 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027/132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVE
 
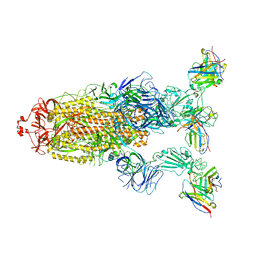 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH027 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
