8YHF
 
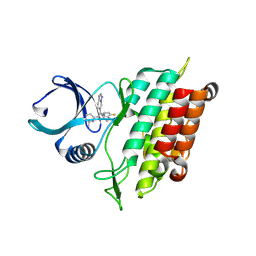 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8YGY
 
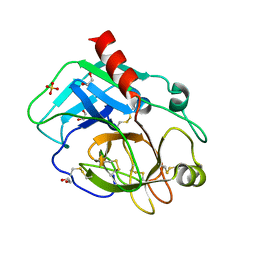 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
8YHS
 
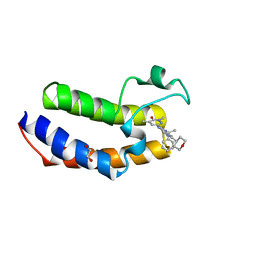 | | The Crystal Structure of BRDT from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dimethyl-6-[6-(oxan-4-yl)-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]pyridazin-3-one, Bromodomain testis-specific protein | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of BRDT from Biortus.
To Be Published
|
|
3RGX
 
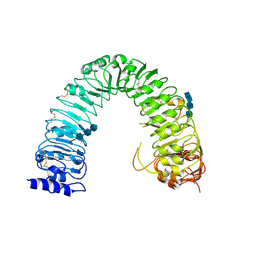 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W, Wang, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
3RGZ
 
 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
6M31
 
 | |
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
6K7P
 
 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
4IPN
 
 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4K6N
 
 | | Crystal structure of yeast 4-amino-4-deoxychorismate lyase | | Descriptor: | Aminodeoxychorismate lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dai, Y.-N, Chi, C.-B, Zhou, K, Cheng, W, Jiang, Y.-L, Ren, Y.-M, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-10 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of yeast 4-amino-4-deoxychorismate lyase
J.Biol.Chem., 288, 2013
|
|
7XQK
 
 | | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, G1/S-specific cyclin-E1, GLYCEROL, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus.
To Be Published
|
|
7Y40
 
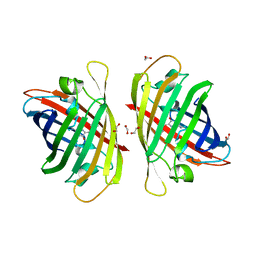 | | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7YAO
 
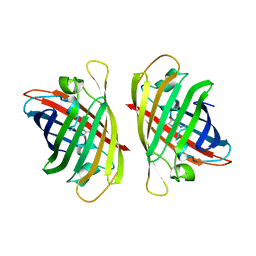 | |
7YRE
 
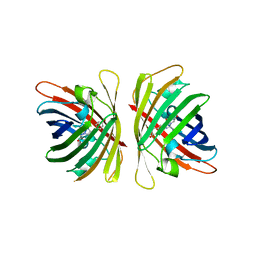 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7ESF
 
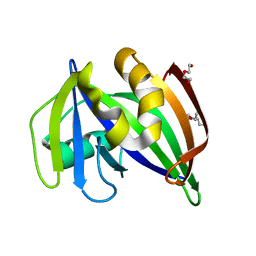 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7ESE
 
 | | The Crystal Structure of human DHFR from Biortus | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, F, Cheng, W, Xu, C, Qi, J, Bao, X, Miao, Q. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of human DHFR from Biortus
To Be Published
|
|
5XUO
 
 | |
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YEU
 
 | | Structural and mechanistic analyses reveal a unique Cas4-like protein in the mimivirus virophage resistance element system | | Descriptor: | MAGNESIUM ION, Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
5YET
 
 | | Structure of R354_WT | | Descriptor: | Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
5YJD
 
 | |
