2I75
 
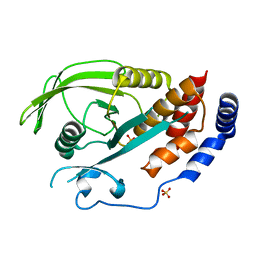 | | Crystal Structure of Human Protein Tyrosine Phosphatase N4 (PTPN4) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Burgess, N, Das, S, Turnbull, A, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2OUO
 
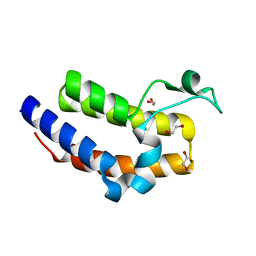 | | Crystal Structure of the Bromo domain 2 in human Bromodomain Containing Protein 4 (BRD4) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Keates, T, Savitsky, P, Burgess, N, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2OSS
 
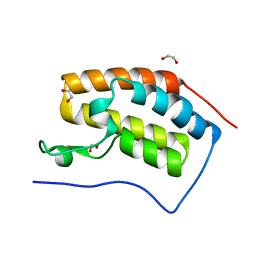 | | Crystal structure of the Bromo domain 1 in human Bromodomain Containing Protein 4 (BRD4) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Keates, T, Savitsky, P, Burgess, N, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2NXB
 
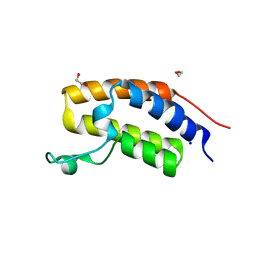 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, SODIUM ION | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, K, Berridge, G, Pike, A, Bunkoczi, G, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5F3I
 
 | | Crystal structure of human KDM4A in complex with compound 54j | | Descriptor: | 8-[4-[2-[4-[3,5-bis(chloranyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F37
 
 | | Crystal structure of human KDM4A in complex with compound 58 | | Descriptor: | 1,2-ETHANEDIOL, 3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F2W
 
 | | Crystal structure of human KDM4A in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F39
 
 | | Crystal structure of human KDM4A in complex with compound 37 | | Descriptor: | 8-(1,3-thiazol-4-yl)-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Lysine-specific demethylase 4A, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F2S
 
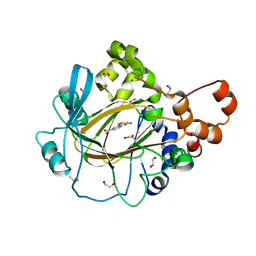 | | Crystal structure of human KDM4A in complex with compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F32
 
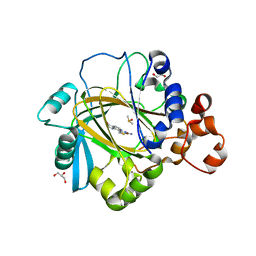 | | Crystal structure of human KDM4A in complex with compound 40 | | Descriptor: | 1,2-ETHANEDIOL, 8-(2-azanyl-1,3-thiazol-4-yl)-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2OO1
 
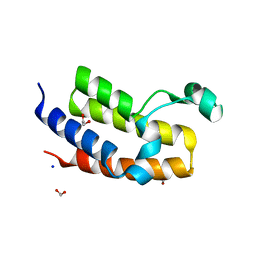 | | Crystal structure of the Bromo domain 2 of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Bromodomain-containing protein 3, ... | | Authors: | Filippakopoulos, P, Bullock, A, Papagrigoriou, E, Keates, T, Cooper, C, Smee, C, Ugochukwu, E, Debreczeni, J, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
6YVS
 
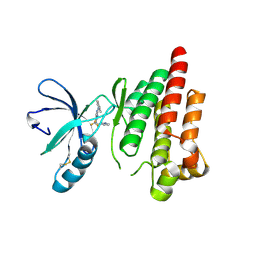 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | Descriptor: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
6YXV
 
 | |
2RFJ
 
 | | Crystal structure of the bromo domain 1 in human bromodomain containing protein, testis specific (BRDT) | | Descriptor: | Bromodomain testis-specific protein | | Authors: | Filippakopoulos, P, Salah, E, Savitsky, P, Keates, T, Parizotto, E, Elkins, J, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7A4Q
 
 | | The Crystal structure of RO4613269 bound to CK2alpha | | Descriptor: | 2-methoxyimino-5-(quinolin-6-ylmethyl)-1,3-thiazol-4-one, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-08-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 2023
|
|
7B3A
 
 | | Crystal structure of PamZ | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Loll, B, Dang, T, Mainz, A, Suessmuth, R, Wahl, M.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Molecular basis of antibiotic self-resistance in a bee larvae pathogen.
Nat Commun, 13, 2022
|
|
6GQE
 
 | |
5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2PA5
 
 | | Crystal structure of human protein tyrosine phosphatase PTPN9 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Pike, A.C.W, Savitsky, P, Papagrigoriou, E, Turnbull, A, Uppenberg, J, Bunkoczi, G, Salah, E, Das, S, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5F3E
 
 | | Crystal structure of human KDM4A in complex with compound 54a | | Descriptor: | 8-[4-[2-[4-(4-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F3C
 
 | | Crystal structure of human KDM4A in complex with compound 52d | | Descriptor: | 8-[4-[2-[(4-fluorophenyl)methyl-methyl-amino]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F3G
 
 | | Crystal structure of human KDM4A in complex with compound 53a | | Descriptor: | 8-[4-[2-[4-[(4-chlorophenyl)methyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5I
 
 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | Descriptor: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2J2I
 
 | | Crystal Structure of the humab PIM1 in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1, SULFATE ION | | Authors: | Debreczeni, J.E, Bullock, A.N, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
