2IVJ
 
 | | Isopenicillin N Synthase From Aspergillus Nidulans (Anaerobic Ac- cyclopropylglycine Fe Complex) | | Descriptor: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-D-CYCLOPROPYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Howard-Jones, A.R, Elkins, J.M, Clifton, I.J, Roach, P.L, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Interactions of Isopenicillin N Synthase with Cyclopropyl-Containing Substrate Analogues Reveal New Mechanistic Insight.
Biochemistry, 46, 2007
|
|
2IVI
 
 | | Isopenicillin N Synthase From Aspergillus Nidulans (Anaerobic Ac- methyl-cyclopropylglycine Fe Complex) | | Descriptor: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-B-METHYL-D-CYCLOPROPYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Elkins, J.M, Howard-Jones, A.R, Clifton, I.J, Roach, P.L, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactions of Isopenicillin N Synthase with Cyclopropyl-Containing Substrate Analogues Reveal New Mechanistic Insight.
Biochemistry, 46, 2007
|
|
1OC1
 
 | | ISOPENICILLIN N SYNTHASE aminoadipoyl-cysteinyl-aminobutyrate-FE COMPLEX | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Long, A.J, Clifton, I.J, Roach, P.L, Baldwin, J.E, Schofield, C.J, Rutledge, P.J. | | Deposit date: | 2003-02-03 | | Release date: | 2004-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Substrate Analogue Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D-Alpha-Aminobutyrate
Biochem.J., 372, 2003
|
|
1OBN
 
 | | ISOPENICILLIN N SYNTHASE aminoadipoyl-cysteinyl-aminobutyrate-FE-NO COMPLEX | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Roach, P.L, Baldwin, J.E, Schofield, C.J, Rutledge, P.J. | | Deposit date: | 2003-01-31 | | Release date: | 2004-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Substrate Analogue Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D-Alpha-Aminobutyrate.
Biochem.J., 372, 2003
|
|
5LLF
 
 | |
5LLB
 
 | |
5LL0
 
 | |
6OLJ
 
 | |
6P9E
 
 | | Crystal structure of IL-36gamma complexed to A-552 | | Descriptor: | (2S)-2-{[4-(3-amino-4-methylphenyl)-6-methylpyrimidin-2-yl]oxy}-3-methoxy-3,3-diphenylpropanoic acid, Interleukin-36 gamma | | Authors: | Argiriadi, M.A, Todorovic, T, Su, Z, Putman, B, Kakavas, S.J, Salte, K.M, McDonald, H.A, Wetter, J.B, Paulsboe, S.E, Sun, Q, Gerstein, C.E, Medina, L, Sielaff, B, Sadhukhan, R, Stockmann, H, Richardson, P.L, Qiu, W, Henry, R.F, Herold, J.M, Shotwell, J.B, McGaraughty, S.P, Honore, P, Gopalakrishnan, S.M, Sun, C.C, Scott, V.E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule IL-36 gamma Antagonist as a Novel Therapeutic Approach for Plaque Psoriasis.
Sci Rep, 9, 2019
|
|
6OK2
 
 | |
6OLL
 
 | |
6FY2
 
 | | Crystal structure of a V2p-reactive RV144 vaccine-like antibody, CAP228-16H, in complex with a heterologous CAP225 V1V2 | | Descriptor: | CAP225 Scaffolded V1V2, CAP228-16H Heavy Chain, CAP228-16H Light Chain, ... | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
6OJZ
 
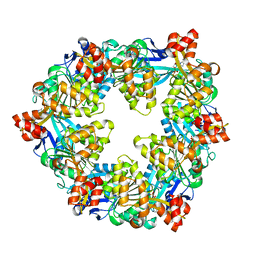 | |
7ZUI
 
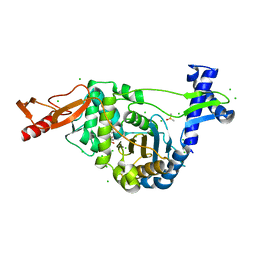 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 5Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUK
 
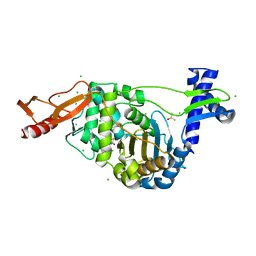 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUJ
 
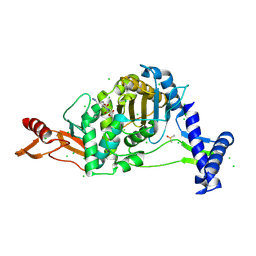 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUH
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUL
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with 8Az lactone - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
6FY3
 
 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-3D, bound to a heterologous V2 peptide | | Descriptor: | CAP228-3D Heavy Chain, CAP228-3D Light Chain, CAP45 V2 peptide | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
7BGJ
 
 | | C. thermophilum Pyruvate Dehydrogenase Complex Core | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Tueting, C, Kastritis, P.L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts.
Cell Rep, 34, 2021
|
|
6FY0
 
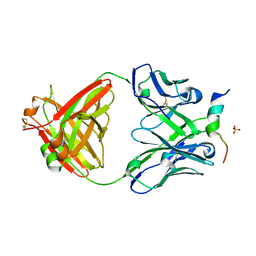 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-16H, bound to a heterologous V2 peptide | | Descriptor: | CAP228-16H Heavy Chain, CAP228-16H Light Chain, CAP45 V2 peptide, ... | | Authors: | Wibmer, C.K, Fernandes, M, Vijayakumar, B, Dirr, H.W, Sayed, Y, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | V2-Directed Vaccine-like Antibodies from HIV-1 Infection Identify an Additional K169-Binding Light Chain Motif with Broad ADCC Activity.
Cell Rep, 25, 2018
|
|
6FY1
 
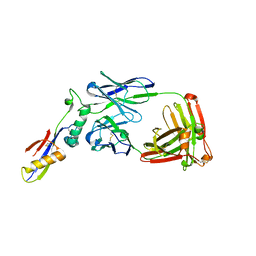 | | Crystal structure of a V2p-reactive RV144 vaccine-like antibody, CAP228-16H, in complex with a scaffolded autologous V1V2 | | Descriptor: | CAP228 Autologous Scaffolded V1V2, CAP228-16H Heavy Chain, CAP228-16H Light Chain | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | Descriptor: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | Authors: | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
3MSI
 
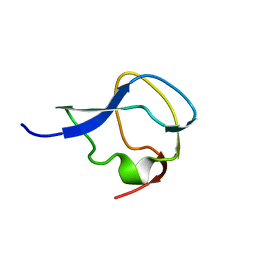 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
3O4V
 
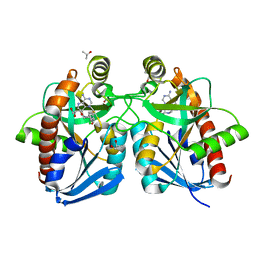 | |
