1HDK
 
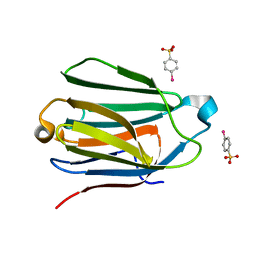 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
1HFZ
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Pike, A.C.W, Brew, K, Acharya, K.R. | | Deposit date: | 1996-06-13 | | Release date: | 1997-07-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase.
Structure, 4, 1996
|
|
1HFX
 
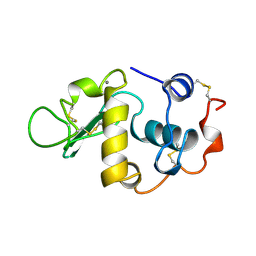 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Pike, A.C.W, Brew, K, Acharya, K.R. | | Deposit date: | 1996-06-13 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase.
Structure, 4, 1996
|
|
1HML
 
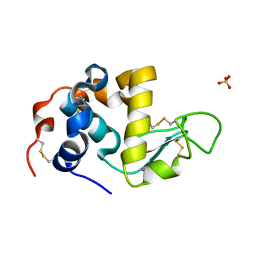 | | ALPHA_LACTALBUMIN POSSESSES A DISTINCT ZINC BINDING SITE | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION, SULFATE ION, ... | | Authors: | Ren, J, Stuart, D.I, Acharya, K.R. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alpha-lactalbumin possesses a distinct zinc binding site.
J.Biol.Chem., 268, 1993
|
|
1O7Y
 
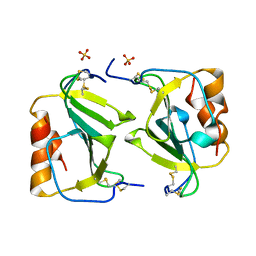 | | Crystal structure of IP-10 M-form | | Descriptor: | SMALL INDUCIBLE CYTOKINE B10, SULFATE ION | | Authors: | Swaminathan, G.J, Holloway, D.E, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-11-20 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Oligomeric Forms of the Ip-10/Cxcl10 Chemokine
Structure, 11, 2003
|
|
1OJQ
 
 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1OJZ
 
 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1QMT
 
 | |
1TTM
 
 | | Human carbonic anhydrase II complexed with 667-coumate | | Descriptor: | 6-OXO-8,9,10,11-TETRAHYDRO-7H-CYCLOHEPTA[C][1]BENZOPYRAN-3-O-SULFAMATE, Carbonic anhydrase II, ZINC ION | | Authors: | Lloyd, M.D, Pederick, R.L, Natesh, R, Woo, L.W.L, Purohit, A, Reed, M.J, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2004-06-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human carbonic anhydrase II at 1.95 A resolution in complex with 667-coumate, a novel anti-cancer agent
Biochem.J., 385, 2005
|
|
6S1Z
 
 | | Crystal structure of Anopheles gambiae AnoACE2 in complex with fosinoprilat | | Descriptor: | Angiotensin-converting enzyme, ZINC ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Cashman, J.S. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of angiotensin-converting enzyme from Anopheles gambiae in its native form and with a bound inhibitor.
Biochem.J., 476, 2019
|
|
6S1Y
 
 | | Crystal structure of Anopheles gambiae AnoACE2 in complex with gamma-Polyglutamic Acid. | | Descriptor: | (2~{S})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Cashman, J.S. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of angiotensin-converting enzyme from Anopheles gambiae in its native form and with a bound inhibitor.
Biochem.J., 476, 2019
|
|
6SH2
 
 | | Crystal structure of human neprilysin E584D in complex with C-type natriuretic peptide. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type natriuretic peptide fragment (CNP), CHLORIDE ION, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of peptide-bound neprilysin reveals key binding interactions.
Febs Lett., 594, 2020
|
|
6SVY
 
 | | Crystal structure of Neprilysin in complex with Sampatrilat-ASP. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, Sampatrilat-Asp, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6SH1
 
 | | Crystal structure of substrate-free human neprilysin E584D. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of peptide-bound neprilysin reveals key binding interactions.
Febs Lett., 594, 2020
|
|
6SUK
 
 | | Crystal structure of Neprilysin in complex with Omapatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-15 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6THY
 
 | | Botulinum neurotoxin A3 Hc domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BoNT/A3, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Febs Open Bio, 10, 2020
|
|
6TT3
 
 | |
6TT1
 
 | |
6TT4
 
 | |
6TWO
 
 | | Binding domain of BoNT/A6 | | Descriptor: | Bont/A1, CHLORIDE ION | | Authors: | Davies, J.R, Britton, A, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
6TWP
 
 | | Binding domain of BoNT/A5 | | Descriptor: | Botulinum neurotoxin A5, CHLORIDE ION | | Authors: | Davies, J.R, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
3ZBV
 
 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
3ZQZ
 
 | | CRYSTAL STRUCTURE OF ANCE IN COMPLEX WITH A SELENIUM ANALOGUE OF CAPTOPRIL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, SELENO-CAPTOPRIL, ... | | Authors: | Akif, M, Masuyer, G, Schwager, S.L.U, Bhuyan, B.J, Mugesh, G, Isaac, R.E, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Angiotensin I- Converting Enzyme in Complex with a Selenium Analogue of Captopril.
FEBS J., 278, 2011
|
|
3ZUQ
 
 | | Crystal structure of an engineered botulinum neurotoxin type B - derivative, LC-B-GS-Hn-B | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZBW
 
 | | Crystal Structure of murine Angiogenin-3 | | Descriptor: | ACETIC ACID, ANGIOGENIN-3, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
